6TQ7
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-334867 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-(2-methyl-1,3-benzoxazol-6-yl)-3-(1,5-naphthyridin-4-yl)urea, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6636 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TOT
 
 | | Crystal structure of the Orexin-1 receptor in complex with lemborexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TP3
 
 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5WPU
 
 | | Crystal structure HpiC1 Y101S | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
3R94
 
 | | AKR1C3 complex with flurbiprofen | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, ... | | Authors: | Yosaatmadja, Y, Teague, R.M, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2011-03-24 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
1F09
 
 | | CRYSTAL STRUCTURE OF THE GREEN FLUORESCENT PROTEIN (GFP) VARIANT YFP-H148Q WITH TWO BOUND IODIDES | | Descriptor: | GREEN FLUORESCENT PROTEIN, IODIDE ION | | Authors: | Wachter, R.M, Yarbrough, D, Kallio, K, Remington, S.J. | | Deposit date: | 2000-05-15 | | Release date: | 2000-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallographic and energetic analysis of binding of selected anions to the yellow variants of green fluorescent protein.
J.Mol.Biol., 301, 2000
|
|
6ZC5
 
 | | Human Adenovirus serotype D10 FiberKnob protein | | Descriptor: | Fiber | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a low-seroprevalence, alpha v beta 6 integrin-selective virotherapy based on human adenovirus type 10.
Mol Ther Oncolytics, 25, 2022
|
|
6ZHM
 
 | | Crystal Structure of Staphylococcus aureus RsgA bound to GDP. | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
5WPS
 
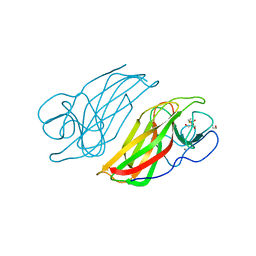 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6ZHL
 
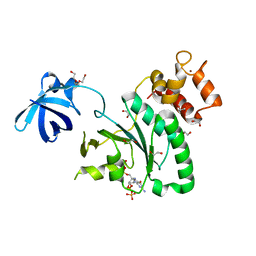 | | Crystal Structure of Staphylococcus aureus RsgA bound to ppGpp. | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
6ZJO
 
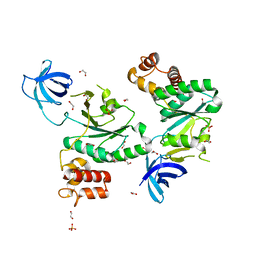 | | Crystal Structure of Staphylococcus aureus RsgA. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
1XTK
 
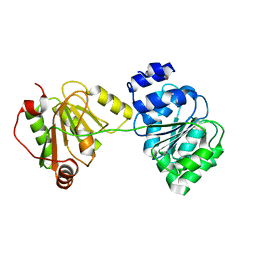 | | structure of DECD to DEAD mutation of human UAP56 | | Descriptor: | BETA-MERCAPTOETHANOL, Probable ATP-dependent RNA helicase p47 | | Authors: | Shi, H, Cordin, O, Minder, C.M, Linder, P, Xu, R.M. | | Deposit date: | 2004-10-22 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human ATP-dependent splicing and export factor UAP56
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XUH
 
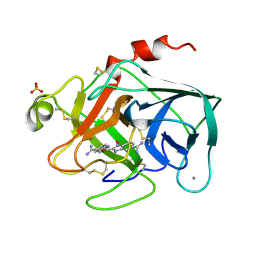 | | TRYPSIN-KETO-BABIM-CO+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-2-BENZIMIDAZOLYL)METHANONE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
6ZG9
 
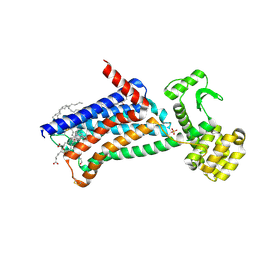 | | Structure of M1-StaR-T4L in complex with GSK1034702 at 2.5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 7-fluoranyl-5-methyl-3-[1-(oxan-4-yl)piperidin-4-yl]-1~{H}-benzimidazol-2-one, Muscarinic acetylcholine receptor M1,Endolysin,Muscarinic acetylcholine receptor M1, ... | | Authors: | Rucktooa, P, Cooke, R.M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From structure to clinic: Design of a muscarinic M1 receptor agonist with potential to treatment of Alzheimer's disease.
Cell, 184, 2021
|
|
6ZFZ
 
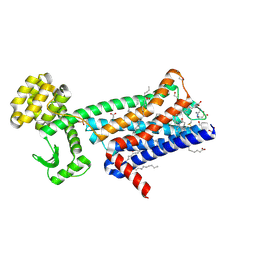 | | Structure of M1-StaR-T4L in complex with 77-LH-28-1 at 2.17A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[3-(4-butylpiperidin-1-yl)propyl]-3,4-dihydroquinolin-2-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rucktooa, P, Cooke, R.M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | From structure to clinic: Design of a muscarinic M1 receptor agonist with potential to treatment of Alzheimer's disease.
Cell, 184, 2021
|
|
4V5Q
 
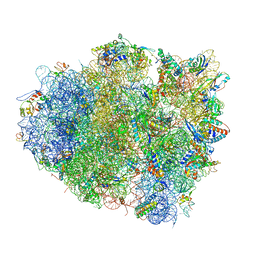 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6ZG4
 
 | | Structure of M1-StaR-T4L in complex with HTL0009936 at 2.35A | | Descriptor: | Muscarinic acetylcholine receptor M1,Endolysin,Muscarinic acetylcholine receptor M1, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Rucktooa, P, Cooke, R.M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | From structure to clinic: Design of a muscarinic M1 receptor agonist with potential to treatment of Alzheimer's disease.
Cell, 184, 2021
|
|
1F0B
 
 | |
4V5E
 
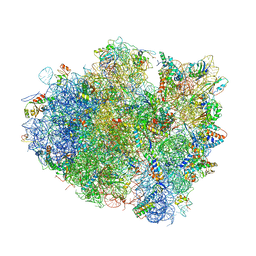 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
1XTI
 
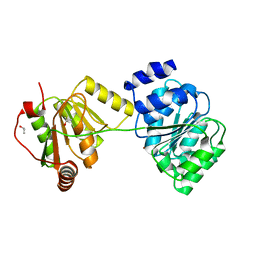 | | Structure of Wildtype human UAP56 | | Descriptor: | ISOPROPYL ALCOHOL, Probable ATP-dependent RNA helicase p47 | | Authors: | Shi, H, Cordin, O, Minder, C.M, Linder, P, Xu, R.M. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the human ATP-dependent splicing and export factor UAP56
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4W4T
 
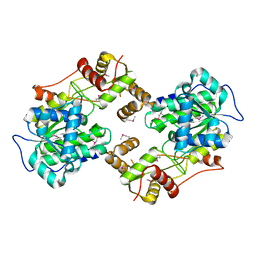 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
3RVN
 
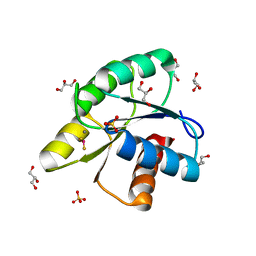 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89Y | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
8G6I
 
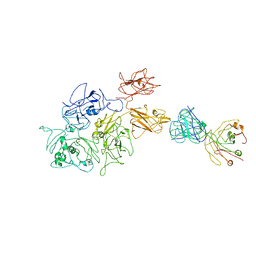 | | Coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VIII chimera from human and pig, NB33 heavy chain, ... | | Authors: | Childers, K.C, Davulcu, O, Haynes, R.M, Lollar, P, Doering, C.B, Coxon, C.H, Spiegel, P.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor.
Blood, 142, 2023
|
|
1XUK
 
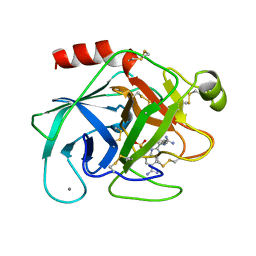 | | TRYPSIN-BABIM-SULFATE, PH 5.9 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE, CALCIUM ION, SULFATE ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
4V5D
 
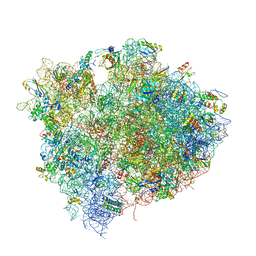 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
