8ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
5OQE
 
 | |
5OQ9
 
 | |
7R7G
 
 | |
6Y5B
 
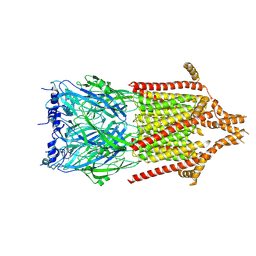 | | 5-HT3A receptor in Salipro (apo, asymmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
7R7F
 
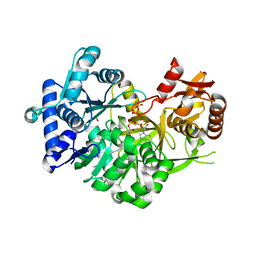 | |
7R7E
 
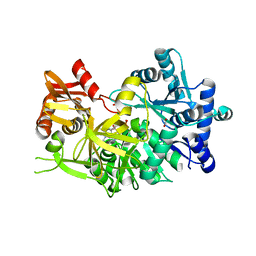 | |
7R36
 
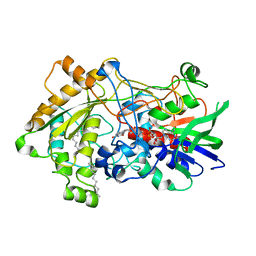 | | Difference-refined structure of fatty acid photodecarboxylase 2 microsecond following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R33
 
 | | Difference-refined structure of fatty acid photodecarboxylase 20 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R34
 
 | | Difference-refined structure of fatty acid photodecarboxylase 900 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R35
 
 | | Difference-refined structure of fatty acid photodecarboxylase 300 ns following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, W. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8AYB
 
 | | anammox-specific FabZ from Scalindua brodae | | Descriptor: | Beta-hydroxyacyl-(Acyl-carrier-protein) dehydratase | | Authors: | Dietl, A, Barends, T.R.M. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of an unusual 3-hydroxyacyl dehydratase (FabZ) from a ladderane-producing organism with an unexpected substrate preference.
J.Biol.Chem., 299, 2023
|
|
7RDO
 
 | | Crystal structure of human galectin-3 CRD in complex with diselenodigalactoside | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]diselanyl}oxane-3,4,5-triol (non-preferred name), CHLORIDE ION, Galectin-3, ... | | Authors: | Kishor, C, Go, R.M, Blanchard, H. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Investigation of the Molecular Details of the Interactions of Selenoglycosides and Human Galectin-3.
Int J Mol Sci, 23, 2022
|
|
7RDP
 
 | |
7B2M
 
 | | Cryo-EM structure of complement C4b in complex with nanobody E3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-C4b nanobody E3, Complement C4 alpha chain, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, van den Bos, R.M, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
8ODW
 
 | | Crystal structure of LbmA Ox-ACP didomain in complex with NADP and ethyl glycinate from the lobatamide PKS (Gynuella sunshinyii) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Francois, R.M.M, Fraley, A.E, Piel, J, Weissman, K.J, Gruez, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Modular Oxime Formation by a trans-AT Polyketide Synthase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OGH
 
 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis with butylacetylphosphonate (BAP) bound | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose-5-phosphate synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gawriljuk, O.V, Oerlemans, R, Groves, R.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Mycobacterium tuberculosis 1-Deoxy-D-Xylulose 5-Phosphate Synthase in Complex with Butylacetylphosphonate
Crystals, 13, 2023
|
|
8OFD
 
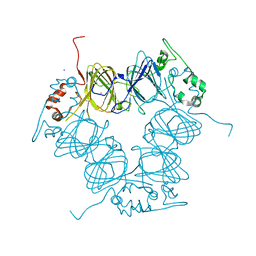 | |
7ROX
 
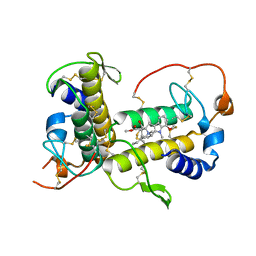 | | BthTX-I complexed with inhibitor MMV | | Descriptor: | 12-methoxy-Nb-methylvoachalotine, Basic phospholipase A2 homolog bothropstoxin-I | | Authors: | Borges, R.J, De Marino, I, Uson, I, Fontes, M.R.M. | | Deposit date: | 2021-08-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of a snake venom phospholipase A 2 -like protein complexed to an inhibitor from Tabernaemontana catharinensis.
Biochimie, 206, 2023
|
|
6ZH7
 
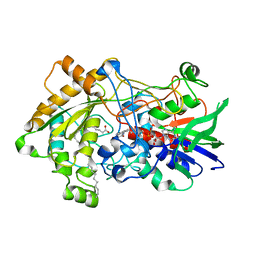 | | Crystal structure of fatty acid photodecarboxylase in the dark state determined by serial femtosecond crystallography at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Weik, M, Schlichting, I, Barends, T.R.M, Colletier, J.P. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8YJM
 
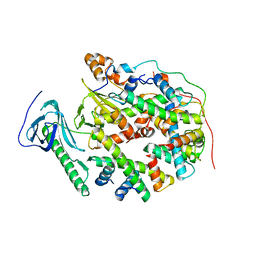 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and a single chain H2B-H2A chimera | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2B 1/2/3/4/6,Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
8YJF
 
 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and an H2A-H2B dimer | | Descriptor: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2A type 1-D, ... | | Authors: | Gan, S.L, Yang, W.S, Xu, R.M. | | Deposit date: | 2024-03-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
5OJE
 
 | | Endothiapepsin with Ligand VSK-B24 | | Descriptor: | (2~{S})-2-azanyl-3-(1~{H}-indol-3-yl)-~{N}-[2-(2,4,6-trimethylphenyl)ethyl]propanamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Gierse, R.M, Magari, F, Groves, M.R, Heine, A, Klebe, G, Hirsch, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-09-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Design and Synthesis of Bioisosteres of Acylhydrazones as Stable Inhibitors of the Aspartic Protease Endothiapepsin.
ChemMedChem, 13, 2018
|
|
5OO0
 
 | | Cdk2(WT) covalent adduct with D28 at C177 | | Descriptor: | Cyclin-dependent kinase 2, methyl 4-propanoyl-2,3-dihydroquinoxaline-1-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8P8P
 
 | | Crystal structure of human Histidine Triad Nucleotide-Binding Protein 1 in complex with 5'-O-[(3-Indolyl)-1-Ethyl]Carbamoyl Ethenoadenosine | | Descriptor: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[2-(1~{H}-indol-3-yl)ethyl]carbamate | | Authors: | Dolot, R.M, Dillenburg, M, Wagner, C.R. | | Deposit date: | 2023-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel inhibitors for hHINT1 protein
To Be Published
|
|
