4W89
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with cellotriose | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4QX1
 
 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4W88
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with a xyloglucan oligosaccharide and TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, ... | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W8B
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in complex with XXLG | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION, ... | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W86
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose and TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, ... | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
1BOZ
 
 | | STRUCTURE-BASED DESIGN AND SYNTHESIS OF LIPOPHILIC 2,4-DIAMINO-6-SUBSTITUTED QUINAZOLINES AND THEIR EVALUATION AS INHIBITORS OF DIHYDROFOLATE REDUCTASE AND POTENTIAL ANTITUMOR AGENTS | | Descriptor: | N6-(2,5-DIMETHOXY-BENZYL)-N6-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (DIHYDROFOLATE REDUCTASE) | | Authors: | Gangjee, A, Vidwans, A.P, Vasudevan, A, Queener, S.F, Kisliuk, R.L, Cody, V, Li, R, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation as inhibitors of dihydrofolate reductases and potential antitumor agents.
J.Med.Chem., 41, 1998
|
|
1HFP
 
 | | COMPARISON OF TERNARY CRYSTAL COMPLEXES OF HUMAN DIHYDROFOLATE REDUCTASE WITH NADPH AND A CLASSICAL ANTITUMOR FUROPYRIMDINE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of ternary crystal complexes of F31 variants of human dihydrofolate reductase with NADPH and a classical antitumor furopyrimidine.
Anti-Cancer Drug Des., 13, 1998
|
|
4TS2
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, magnesium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4RVY
 
 | | Serial Time resolved crystallography of Photosystem II using a femtosecond X-ray laser. The S state after two flashes (S3) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.-H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-11-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
4WXU
 
 | | Crystal Structure of the Selenomthionine Incorporated Myocilin Olfactomedin Domain E396D Variant. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
1HFQ
 
 | | COMPARISON OF TERNARY CRYSTAL COMPLEXES OF HUMAN DIHYDROFOLATE REDUCTASE WITH NADPH AND A CLASSICAL ANTITUMOR FUROPYRIMDINE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of ternary crystal complexes of F31 variants of human dihydrofolate reductase with NADPH and a classical antitumor furopyrimidine.
Anti-Cancer Drug Des., 13, 1998
|
|
4WXS
 
 | | Crystal Structure of the E396D SNP Variant of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
4TS0
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, barium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, BARIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1HFR
 
 | | COMPARISON OF TERNARY CRYSTAL COMPLEXES OF HUMAN DIHYDROFOLATE REDUCTASE WITH NADPH AND A CLASSICAL ANTITUMOR FUROPYRIMDINE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of ternary crystal complexes of F31 variants of human dihydrofolate reductase with NADPH and a classical antitumor furopyrimidine.
Anti-Cancer Drug Des., 13, 1998
|
|
1HDS
 
 | |
3GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED TRYPANOTHIONE COMPLEX | | Descriptor: | 2-AMINO-4-[4-(4-AMINO-4-CARBOXY-BUTYRYLAMINO)-5,8,19,22-TETRAOXO-1,2-DITHIA-6,9,13,18,21-PENTAAZA-CYCLOTETRACOS-23-YLCARBAMOYL]-BUTYRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
6O7K
 
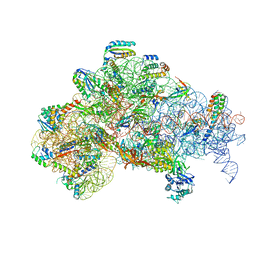 | | 30S initiation complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Frank, J, Gonzalez Jr, R.L, kaledhonkar, S, Fu, Z, Caban, K, Li, W, Chen, B, Sun, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-29 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Late steps in bacterial translation initiation visualized using time-resolved cryo-EM.
Nature, 570, 2019
|
|
6OPA
 
 | |
6OMC
 
 | | capsid of T5 virion | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1TWZ
 
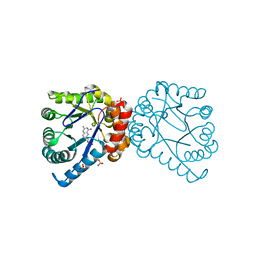 | | Dihydropteroate Synthetase, With Bound Substrate Analogue PtP, From Bacillus anthracis | | Descriptor: | DHPS, Dihydropteroate synthase, PTERIN-6-YL-METHYL-MONOPHOSPHATE, ... | | Authors: | Babaoglu, K, Qi, J, Lee, R.L, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
1TZG
 
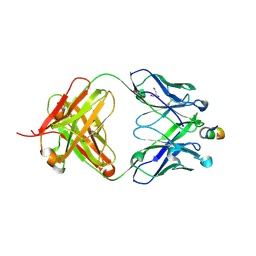 | | Crystal structure of HIV-1 neutralizing human Fab 4E10 in complex with a 13-residue peptide containing the 4E10 epitope on gp41 | | Descriptor: | Envelope polyprotein GP160, Fab 4E10, GLYCEROL | | Authors: | Cardoso, R.M.F, Zwick, M.B, Stanfield, R.L, Kunert, R, Binley, J.M, Katinger, H, Burton, D.R, Wilson, I.A. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broadly Neutralizing Anti-HIV Antibody 4E10 Recognizes a Helical Conformation of a Highly Conserved Fusion-Associated Motif in gp41
Immunity, 22, 2005
|
|
6OMA
 
 | |
1U4S
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedisulphonic acid | | Descriptor: | L-lactate dehydrogenase, NAPHTHALENE-2,6-DISULFONIC ACID | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
