2VAD
 
 | | Monomeric red fluorescent protein, DsRed.M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RED FLUORESCENT PROTEIN, ... | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
1NHK
 
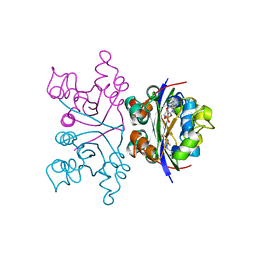 | |
2W99
 
 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
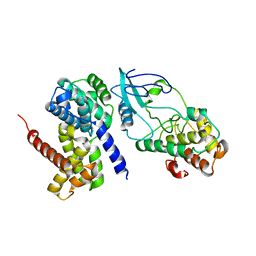
1Q1J
 
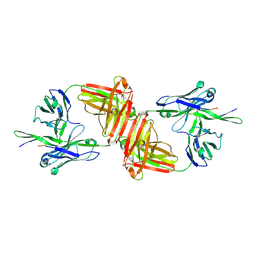 | | Crystal Structure Analysis of anti-HIV-1 Fab 447-52D in complex with V3 peptide | | Descriptor: | Fab 447-52D, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Williams, C, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2003-07-21 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D.
Structure, 12, 2004
|
|
1O5E
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, Serine protease hepsin | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1NSK
 
 | |
1NND
 
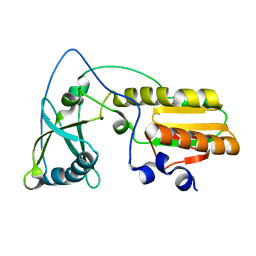 | | Arginine 116 is Essential for Nucleic Acid Recognition by the Fingers Domain of Moloney Murine Leukemia Virus Reverse Transcriptase | | Descriptor: | Reverse Transcriptase | | Authors: | Crowther, R.L, Remeta, D.P, Minetti, C.A, Das, D, Montano, S.P, Georgiadis, M.M. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and energetic characterization of nucleic acid-binding to the fingers domain of Moloney murine leukemia virus reverse transcriptase
Proteins, 57, 2004
|
|
1NWE
 
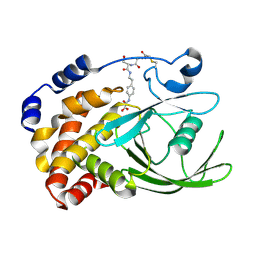 | | Ptp1B R47C Modified at C47 with N-[4-(2-{2-[3-(2-Bromo-acetylamino)-propionylamino]-3-hydroxy-propionylamino}-ethyl)-phenyl]-oxalamic acid | | Descriptor: | N-[4-(2-{2-[3-(2-BROMO-ACETYLAMINO)-PROPIONYLAMINO]-3-HYDROXY-PROPIONYLAMINO}-ETHYL)-PHENYL]-OXALAMIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Erlanson, D.A, McDowell, R.S, He, M.M, Randal, M, Simmons, R.L, Kung, J, Waight, A, Hansen, S.K. | | Deposit date: | 2003-02-06 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of a New Phosphotyrosine Mimetic for PTP1B Using Breakaway Tethering
J.Am.Chem.Soc., 125, 2003
|
|
1O0V
 
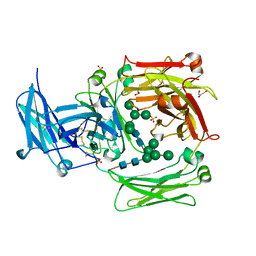 | | The crystal structure of IgE Fc reveals an asymmetrically bent conformation | | Descriptor: | GLYCEROL, Immunoglobulin heavy chain epsilon-1, SULFATE ION, ... | | Authors: | Wan, T, Beavil, R.L, Fabiane, S.M, Beavil, A.J, Sohi, M.K, Keown, M, Young, R.J, Henry, A.J, Owens, R.J, Gould, H.J, Sutton, B.J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of IgE Fc reveals an asymmetrically bent conformation
NAT.IMMUNOL., 3, 2002
|
|
2Y3A
 
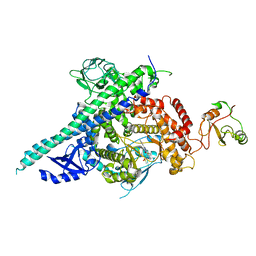 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
1RLW
 
 | |
1OZH
 
 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. | | Descriptor: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1RCY
 
 | | RUSTICYANIN (RC) FROM THIOBACILLUS FERROOXIDANS | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Walter, R.L, Friedman, A.M, Ealick, S.E, Blake II, R.C, Proctor, P, Shoham, M. | | Deposit date: | 1996-04-10 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability.
J.Mol.Biol., 263, 1996
|
|
3M54
 
 | | SET7/9 Y305F in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3MER
 
 | | Crystal Structure of the methyltransferase Slr1183 from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Rossi, P, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR145
To be Published
|
|
3M57
 
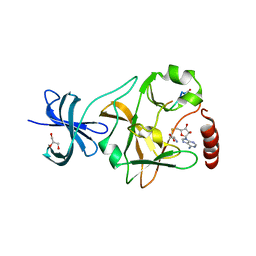 | | SET7/9 Y245A in complex with TAF10 peptide and AdoHcy | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
5ILT
 
 | | Crystal structure of bovine Fab A01 | | Descriptor: | bovine Fab A01 heavy chain, bovine Fab A01 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
5IN9
 
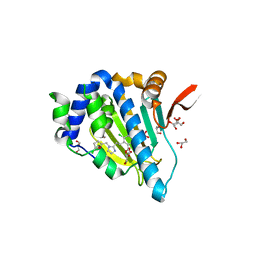 | | Crystal structure of Grp94 bound to methyl 3-chloro-2-(2-(1-((5-chlorofuran-2-yl)methyl)-1H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, an inhibitor based on the BnIm and Radamide scaffolds. | | Descriptor: | Endoplasmin, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Lieberman, R.L, Huard, D.J.E, Kizziah, J.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of Glucose Regulated Protein 94-Selective Inhibitors Based on the BnIm and Radamide Scaffold.
J.Med.Chem., 59, 2016
|
|
6UB8
 
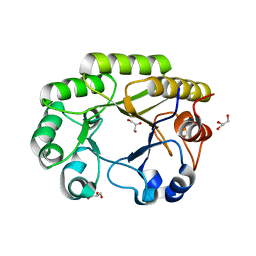 | | Crystal structure of a GH128 (subgroup VI) exo-beta-1,3-glucanase from Aureobasidium namibiae (AnGH128_VI) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein | | Authors: | Santos, C.R, Vieira, P.S, Domingues, M.N, Cordeiro, R.L, Tomazini, A, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
2G8J
 
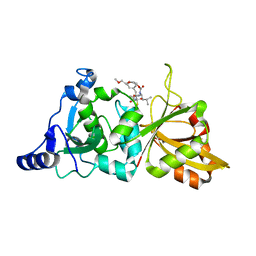 | | Calpain 1 proteolytic core in complex with SNJ-1945, a alpha-ketoamide-type inhibitor. | | Descriptor: | ((1S)-1-((((1S)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2,3-DIOXOPROPYL)AMINO)CARBONYL)-3-METHYLBUTYL)CARBAMIC ACID 5-METHOXY-3-OXAPENTYL ESTER, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
2FSY
 
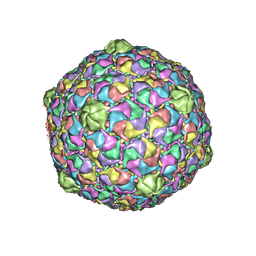 | | Bacteriophage HK97 Pepsin-treated Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
6ULL
 
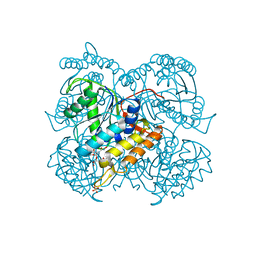 | | BshB from Bacillus subtilis complexed with a substrate analogue | | Descriptor: | (2S)-2-({2-deoxy-2-[(hydroxycarbamoyl)amino]-alpha-D-glucopyranosyl}oxy)butanedioic acid, N-acetyl-alpha-D-glucosaminyl L-malate deacetylase 1, SULFATE ION, ... | | Authors: | Cook, P.D, Castleman, M.M, Woodward, R.L. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray crystallographic structure of BshB, the zinc-dependent deacetylase involved in bacillithiol biosynthesis.
Protein Sci., 29, 2020
|
|
4P4U
 
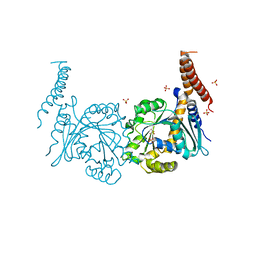 | | Nucleotide-free stalkless-MxA | | Descriptor: | Interferon-induced GTP-binding protein Mx1, SULFATE ION | | Authors: | Rennie, M.L, McKelvie, S.A, Bulloch, E.M, Kingston, R.L. | | Deposit date: | 2014-03-13 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient Dimerization of Human MxA Promotes GTP Hydrolysis, Resulting in a Mechanical Power Stroke.
Structure, 22, 2014
|
|
2FRP
 
 | | Bacteriophage HK97 Expansion Intermediate IV | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
3NZP
 
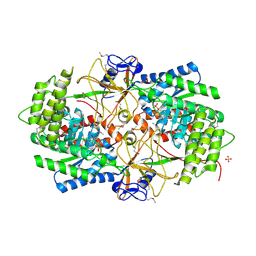 | | Crystal Structure of the Biosynthetic Arginine decarboxylase SpeA from Campylobacter jejuni, Northeast Structural Genomics Consortium Target BR53 | | Descriptor: | Arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
