2MC1
 
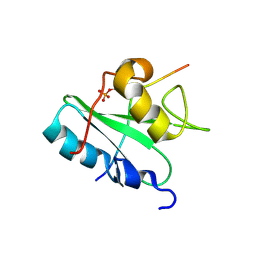 | | Solution structure of the Vav1 SH2 domain complexed with a Syk-derived singly phosphorylated peptide | | Descriptor: | Proto-oncogene vav, Tyrosine-protein kinase SYK | | Authors: | Chen, C, Piraner, D, Gorenstein, N.M, Geahlen, R.L, Post, C.B. | | Deposit date: | 2013-08-13 | | Release date: | 2013-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain.
Biopolymers, 99, 2013
|
|
7SJU
 
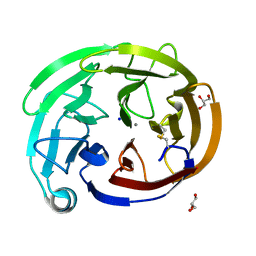 | | Myocilin OLF mutant T293K | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SIJ
 
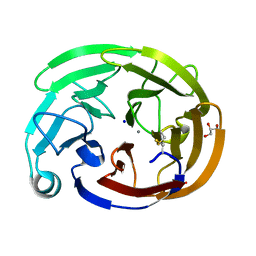 | | Myocilin OLF mutant E352K | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-14 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SJV
 
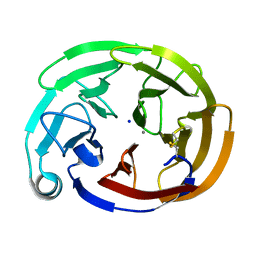 | | Myocilin OLF mutant T353I | | Descriptor: | CALCIUM ION, Myocilin, C-terminal fragment, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SKF
 
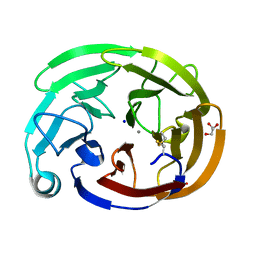 | | Myocilin OLF mutant A445V | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.282 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
2N54
 
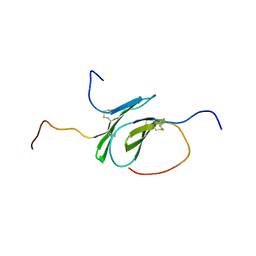 | | Solution structure of a disulfide stabilized XCL1 dimer | | Descriptor: | Lymphotactin | | Authors: | Tyler, R.C, Tuinstra, R.L, Peterson, F.F, Volkman, B.F. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering Metamorphic Chemokine Lymphotactin/XCL1 into the GAG-Binding, HIV-Inhibitory Dimer Conformation.
Acs Chem.Biol., 10, 2015
|
|
7SKE
 
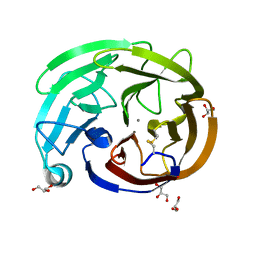 | | Myocilin OLF mutant R296H | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SJT
 
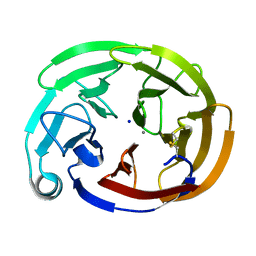 | | Myocilin OLF mutant K398R | | Descriptor: | CALCIUM ION, Myocilin, C-terminal fragment, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SJW
 
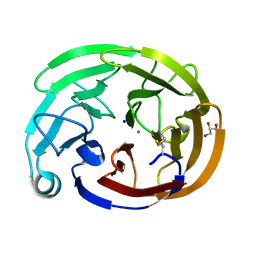 | | Myocilin OLF mutant L303I | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SKG
 
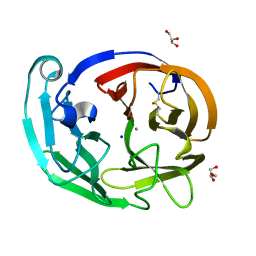 | | Myocilin OLF mutant V329M | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SIB
 
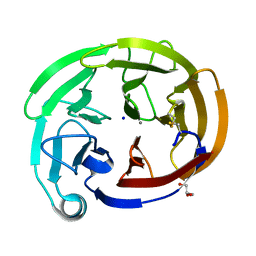 | | Myocilin OLF mutant K500R | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SKD
 
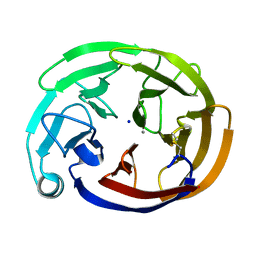 | | Myocilin OLF mutant S331L | | Descriptor: | CALCIUM ION, Myocilin, C-terminal fragment, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
8DYK
 
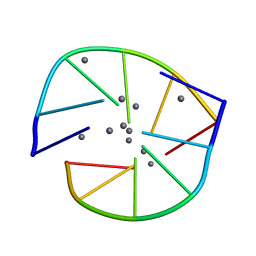 | |
1LMK
 
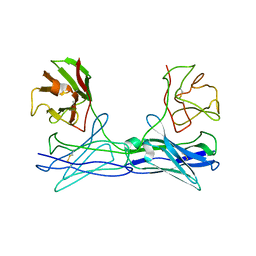 | | THE STRUCTURE OF A BIVALENT DIABODY | | Descriptor: | ANTI-PHOSPHATIDYLINOSITOL SPECIFIC PHOSPHOLIPASE C DIABODY | | Authors: | Williams, R.L. | | Deposit date: | 1994-08-29 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a diabody, a bivalent antibody fragment.
Structure, 2, 1994
|
|
2N9P
 
 | | Solution structure of RNF126 N-terminal zinc finger domain in complex with BAG6 Ubiquitin-like domain | | Descriptor: | E3 ubiquitin-protein ligase RNF126, Large proline-rich protein BAG6, ZINC ION | | Authors: | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
8E5I
 
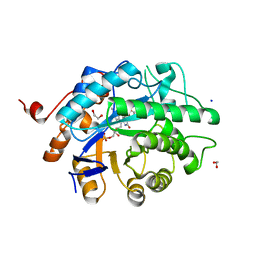 | |
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SXO
 
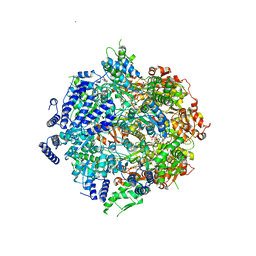 | | Yeast Lon (PIM1) with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Yang, J, Song, A.S, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2022-01-12 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of hexameric yeast Lon protease (PIM1) highlights the importance of conserved structural elements.
J.Biol.Chem., 298, 2022
|
|
1YNV
 
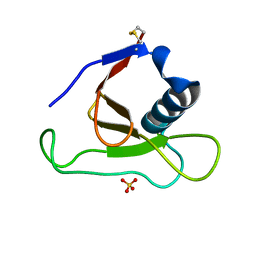 | | Asp79 makes a large, unfavorable contribution to the stability of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Trevino, S.R, Gokulan, K, Newsom, S, Thurlkill, R.L, Shaw, K.L, Mitkevich, V.A, Makarov, A.A, Sacchettini, J.C, Scholtz, J.M, Pace, C.N. | | Deposit date: | 2005-01-25 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Asp79 Makes a Large, Unfavorable Contribution to the Stability of RNase Sa.
J.Mol.Biol., 354, 2005
|
|
5NP0
 
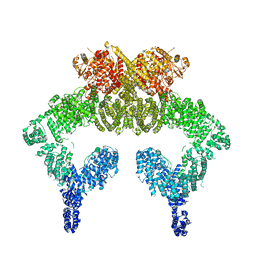 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
7SUF
 
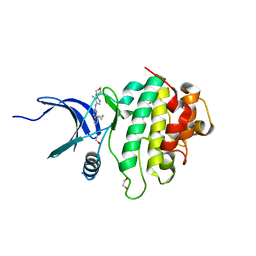 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 06 | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclopropyl-N-[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]quinazolin-2-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
8E5H
 
 | |
7SUJ
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 24 | | Descriptor: | (3R,4R)-4-{4-[6-chloro-2-({1-[(1R)-2,2-difluorocyclopropyl]-5-methyl-1H-pyrazol-4-yl}amino)quinazolin-7-yl]piperidin-1-yl}-4-methyloxolan-3-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
8EDF
 
 | |
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
