9FTH
 
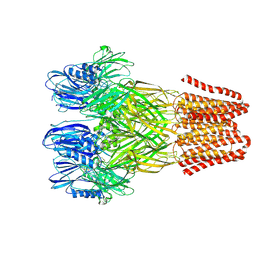 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM Ca2+ | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9FTJ
 
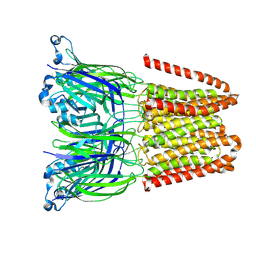 | | Closed and disordered conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
8SAH
 
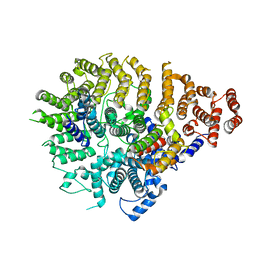 | | Huntingtin C-HEAT domain in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Alteen, M.G, Arrowsmith, C.H, Lea, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-31 | | Release date: | 2023-04-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Delineation of functional subdomains of Huntingtin protein and their interaction with HAP40.
Structure, 31, 2023
|
|
6E0E
 
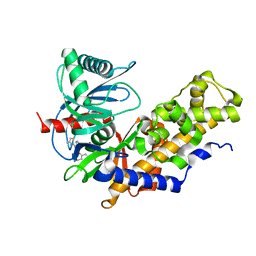 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
3D4K
 
 | | Concanavalin A Complexed to a Synthetic Analog of the Trimannoside | | Descriptor: | CALCIUM ION, Concanavalin-A, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Foley, B.L, Dyekjaer, J.D, Woods, R.J. | | Deposit date: | 2008-05-14 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Involvement of water in carbohydrate-protein binding: concanavalin A revisited.
J.Am.Chem.Soc., 130, 2008
|
|
3IJH
 
 | | Structure of S67-27 in Complex with Ko | | Descriptor: | Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
3IJS
 
 | | Structure of S67-27 in Complex with TSBP | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-alpha-D-glucopyranose, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
3IKC
 
 | | Structure of S67-27 in Complex with Kdo(2.8)-7-O-methyl-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-(1E)-prop-1-en-1-yl 3-deoxy-7-O-methyl-alpha-D-manno-oct-2-ulopyranosidonic acid, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
3D1Q
 
 | | Structure of the PTP-Like Phytase Expressed by Selenomonas Ruminantium at an Ionic Strength of 400 mM | | Descriptor: | CHLORIDE ION, GLYCEROL, Myo-inositol hexaphosphate phosphohydrolase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of ionic strength and oxidation on the P-loop conformation of the protein tyrosine phosphatase-like phytase, PhyAsr.
Febs J., 275, 2008
|
|
9DK4
 
 | | ancestral hydroxynitrile lyase with fifteen substitutions | | Descriptor: | Fifteen-substitution variant of an ancestral hydroxynitrile lyase, SODIUM ION | | Authors: | Sarak, S.C, Tan, P, Evans III, R.L, Shi, K, Kazlauskas, R.J, Aihara, H, Pierce, C.T, Walsh, M.E, Greenberg, L.R. | | Deposit date: | 2024-09-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | To be published
To Be Published
|
|
3IA2
 
 | | Pseudomonas fluorescens esterase complexed to the R-enantiomer of a sulfonate transition state analog | | Descriptor: | (2R)-butane-2-sulfonate, Arylesterase, GLYCEROL, ... | | Authors: | Schrag, J.D, Kazlauskas, R.J, Jiang, Y, Morley, K. | | Deposit date: | 2009-07-13 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Different active-site loop orientation in serine hydrolases versus acyltransferases.
Chembiochem, 12, 2011
|
|
3CL4
 
 | | Crystal structure of bovine coronavirus hemagglutinin-esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4Q4I
 
 | | Crystal structure of E.coli aminopeptidase N in complex with amastatin | | Descriptor: | Amastatin, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
9FVS
 
 | | Crystal structure of Heme-Oxygenase mutant G139A from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING CO, heme oxygenase (biliverdin-producing) | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-06-28 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
9FY4
 
 | | Crystal structure of Heme-Oxygenase mutant I143K from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-07-02 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
3IXY
 
 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
5EYV
 
 | | Crystal Structure of Adenylosuccinate lyase from Schistosoma mansoni in APO form. | | Descriptor: | Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L.E, Nettleship, J.E, Owens, R.J, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
5LQF
 
 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
6EOE
 
 | | Crystal structure of AMPylated GRP78 with nucleotide | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-DIPHOSPHATE, CITRATE ANION, ... | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
1Z4U
 
 | | hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00799585 | | Descriptor: | (2Z)-2-[(1-ADAMANTYLCARBONYL)AMINO]-3-[4-(2-BROMOPHENOXY)PHENYL]PROP-2-ENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M, Nugent, R, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R. | | Deposit date: | 2005-03-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 2: Evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid
Bioorg.Med.Chem.Lett., 15, 2005
|
|
9FW4
 
 | | Crystal structure of Heme-Oxygenase mutant H20C from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING CO, heme oxygenase (biliverdin-producing) | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-06-28 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
6EOB
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 1) | | Descriptor: | 78 kDa glucose-regulated protein, PHOSPHATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
8SGO
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus pregnenolone sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Legesse, D.H, Fan, C, Teng, J, Zhuang, Y, Howard, R.J, Noviello, C.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural insights into opposing actions of neurosteroids on GABA A receptors.
Nat Commun, 14, 2023
|
|
4Q9N
 
 | | Crystal structure of Chlamydia trachomatis enoyl-ACP reductase (FabI) in complex with NADH and AFN-1252 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Yao, J, Abdelrahman, Y, Robertson, R.M, Cox, J.V, Belland, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Type II Fatty Acid Synthesis Is Essential for the Replication of Chlamydia trachomatis.
J.Biol.Chem., 289, 2014
|
|
5FM2
 
 | | Crystal structure of hyper-phosphorylated RET kinase domain with (proximal) juxtamembrane segment | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Barry, R, Borg, A, Orme, M, Mouilleron, S, Martinez-Torres, R.J, Meier, P, McDonald, N.Q. | | Deposit date: | 2015-10-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RET Functions as a Dual-Specificity Kinase that Requires Allosteric Inputs from Juxtamembrane Elements.
Cell Rep, 17, 2016
|
|
