7TZ1
 
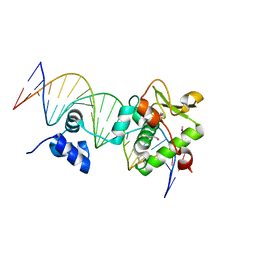 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
1NQG
 
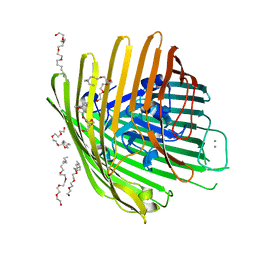 | | OUTER MEMBRANE COBALAMIN TRANSPORTER (BTUB) FROM E. COLI, WITH BOUND CALCIUM | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, vitamin b12 receptor | | Authors: | Chimento, D.P, Mohanty, A.K, Kadner, R.J, Wiener, M.C. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Substrate-induced transmembrane signaling in the cobalamin transporter BtuB
Nat.Struct.Biol., 10, 2003
|
|
6CWM
 
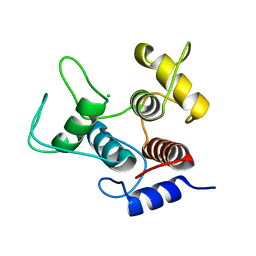 | | Crystal structure of SpaA-SLH/G109A | | Descriptor: | CHLORIDE ION, Surface (S-) layer glycoprotein | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
4PKA
 
 | |
2W2S
 
 | | Structure of the Lagos bat virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
6BR6
 
 | | N2 neuraminidase in complex with a novel antiviral compound | | Descriptor: | (1R)-4-acetamido-3-amino-1,5-anhydro-2,3,4-trideoxy-1-sulfo-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, L.H, Ve, T, Pascolutti, M, Hadhazi, A, Bailly, B, Thomson, R.J, Gao, G.F, von Itzstein, M. | | Deposit date: | 2017-11-30 | | Release date: | 2018-03-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03983879 Å) | | Cite: | A Sulfonozanamivir Analogue Has Potent Anti-influenza Virus Activity.
ChemMedChem, 13, 2018
|
|
6BXA
 
 | | Crystal structure of N-terminal fragment of Zebrafish Toll-Like Receptor 5 (TLR5) with Lamprey Variable Lymphocyte Receptor 2 (VLR2) bound | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gunn, R.J, Wilson, I.A, Cooper, M.D, Herrin, B.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | VLR Recognition of TLR5 Expands the Molecular Characterization of Protein Antigen Binding by Non-Ig-based Antibodies.
J. Mol. Biol., 430, 2018
|
|
4NPR
 
 | | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase GH12 | | Authors: | Cordeiro, R.L, Santos, C.R, Furtado, G.P, Damasio, A.R.L, Polizeli, M.L.T.M, Ward, R.J, Murakami, M.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus
To be Published
|
|
7V0N
 
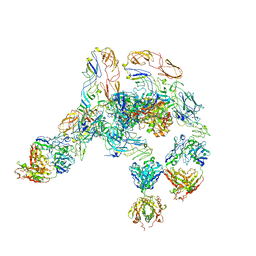 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21 | | Descriptor: | IgG 21 Fab heavy chain, IgG 21 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7V0O
 
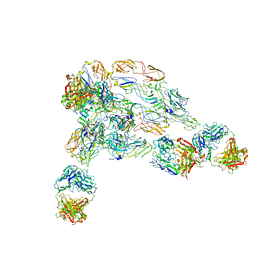 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94 | | Descriptor: | IgG 94 Fab heavy chain, IgG 94 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7V0P
 
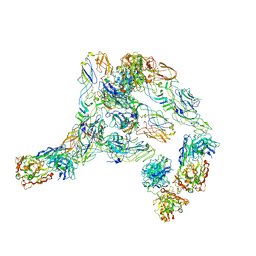 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106 | | Descriptor: | IgG 106 Fab heavy chain, IgG 106 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6COH
 
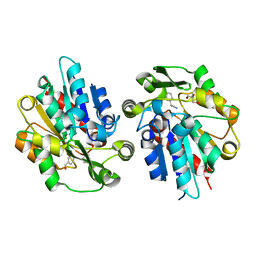 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I with benzaldehyde, MANDELIC ACID NITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, Alpha-hydroxynitrile lyase, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6CE6
 
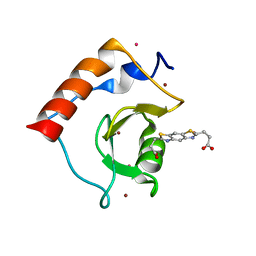 | | Structure of HDAC6 zinc-finger ubiquitin binding domain soaked with 3,3'-(benzo[1,2-d:5,4-d']bis(thiazole)-2,6-diyl)dipropionic acid | | Descriptor: | 3,3'-(benzo[1,2-d:5,4-d']bis[1,3]thiazole-2,6-diyl)dipropanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CWF
 
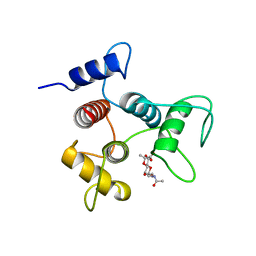 | |
6CWN
 
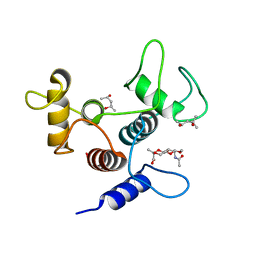 | | Crystal structure of SpaA-SLH/G109A in complex with 4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Surface (S-) layer glycoprotein, methyl 2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
6COF
 
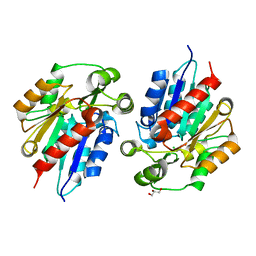 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6CQ3
 
 | |
6COC
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,F179L,A209I with benzaldehyde | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
4OX3
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | PHOSPHATE ION, Putative carboxypeptidase YodJ, ZINC ION | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
1NQF
 
 | | OUTER MEMBRANE COBALAMIN TRANSPORTER (BTUB) FROM E. COLI, METHIONINE SUBSTIUTION CONSTRUCT FOR SE-MET SAD PHASING | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, Vitamin B12 receptor | | Authors: | Chimento, D.P, Mohanty, A.K, Kadner, R.J, Wiener, M.C. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate-induced transmembrane signaling in the cobalamin transporter BtuB
Nat.Struct.Biol., 10, 2003
|
|
6DGM
 
 | | Streptococcus pyogenes phosphoglycerol transferase GacH in complex with sn-glycerol-1-phosphate | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Phosphoglycerol transferase GacH, ... | | Authors: | Edgar, R.J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-05-17 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides.
Nat.Chem.Biol., 15, 2019
|
|
6DXH
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-tert-butylphenyl)-4-oxobutanoate | | Descriptor: | 4-(4-tert-butylphenyl)-4-oxobutanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Harding, R.J, Mann, M.K, Ravichandran, M, Ferreira de Freitas, R, Franzoni, I, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
6DXT
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 3-(5-phenyl-1,3,4-oxadiazol-2-yl)propanoate | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-phenyl-1,3,4-oxadiazol-2-yl)propanoic acid, UNKNOWN ATOM OR ION, ... | | Authors: | Mann, M.K, Harding, R.J, Ravichandran, M, Ferreira de Freitas, R, Franzoni, I, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
4PPJ
 
 | | Crystal structure of Phanta, a weakly fluorescent photochromic GFP-like protein. ON state | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
6EOC
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 2) | | Descriptor: | 78 kDa glucose-regulated protein, CITRATE ANION, SULFATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
