6BBS
 
 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with brinzolamide | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, Aggarwal, M, McKenna, R. | | Deposit date: | 2017-10-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
2KQ2
 
 | | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A | | Descriptor: | Ribonuclease H-related protein | | Authors: | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-24 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A
To be Published
|
|
6B03
 
 | |
3C49
 
 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor KU0058948 | | Descriptor: | 4-[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorobenzyl]phthalazin-1(2H)-one, Poly(ADP-ribose) polymerase 3 | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Helleday, T, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
3C4H
 
 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor DR2313 | | Descriptor: | 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Poly(ADP-ribose) polymerase 3 | | Authors: | Lehtio, L, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
4FCR
 
 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 2-{[4-(2-chloro-4,5-dimethoxyphenyl)-5-cyano-7H-pyrrolo[2,3-d]pyrimidin-2-yl]sulfanyl}-N,N-dimethylacetamide, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
3GUW
 
 | | Crystal Structure of the TatD-like Protein (AF1765) from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR121 | | Descriptor: | ZINC ION, uncharacterized protein AF_1765 | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR121
To be Published
|
|
2KRT
 
 | | Solution NMR Structure of a Conserved Hypothetical Membrane Lipoprotein Obtained from Ureaplasma parvum: Northeast Structural Genomics Consortium Target UuR17A (139-239) | | Descriptor: | Conserved hypothetical membrane lipoprotein | | Authors: | Mani, R, Swapna, G, Janjua, H, Ciccosanti, C, Huang, Y, Patel, D, Xiao, R, Acton, T, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Conserved Hypothetical Membrane Lipoprotein obtained from Ureaplasma parvum : Northeast Structural Genomics Consortium Target UuR17A (139-239)
To be Published
|
|
3GYG
 
 | | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis | | Descriptor: | MAGNESIUM ION, NTD biosynthesis operon putative hydrolase ntdB | | Authors: | Nocek, B, Stein, A, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis
To be Published
|
|
4OO7
 
 | | THE 1.55A CRYSTAL STRUCTURE of NAF1 (MINER1): THE REDOX-ACTIVE 2FE-2S PROTEIN | | Descriptor: | CDGSH iron-sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Tamir, S, Eisenberg-Domovich, Y, Colman, A.R, Stofleth, J.T, Lipper, C.H, Paddock, M.L, Jenning, P.A, Livnah, O, Nechushtai, R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OOK
 
 | | Third Metal bound M.tuberculosis methionine aminopeptidase | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 2, SODIUM ION | | Authors: | Reddi, R, Addlagatta, A. | | Deposit date: | 2014-02-03 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
6DPU
 
 | | Undecorated GMPCPP microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Separating the effects of nucleotide and EB binding on microtubule structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DR8
 
 | | Metallo-beta-lactamase from Cronobacter sakazakii (Enterobacter sakazakii) HARLDQ motif mutant S60/R118H/Q121H/K254H | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6D9W
 
 | | Crystal structure of Deinococcus radiodurans MntH, an Nramp-family transition metal transporter, in the inward-open apo state | | Descriptor: | Divalent metal cation transporter MntH, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Gaudet, R, Bane, L.B, Weihofen, W.A, Singharoy, A, Zimanyi, C.M, Bozzi, A.T. | | Deposit date: | 2018-04-30 | | Release date: | 2019-02-13 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.941 Å) | | Cite: | Structures in multiple conformations reveal distinct transition metal and proton pathways in an Nramp transporter.
Elife, 8, 2019
|
|
6D66
 
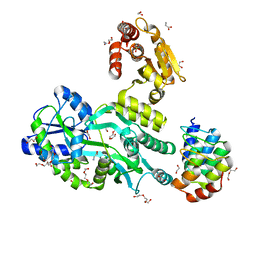 | | Crystal structure of the human dual specificity 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, D-ALANINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gumpena, R, Waugh, D.S, Lountos, G.T. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6DFU
 
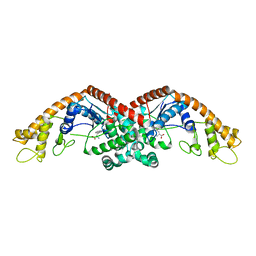 | | Tryptophan--tRNA ligase from Haemophilus influenzae. | | Descriptor: | TRYPTOPHAN, Tryptophan--tRNA ligase | | Authors: | Osipiuk, J, Maltseva, N, Mulligan, R, Grimshaw, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tryptophan--tRNA ligase from Haemophilus influenzae.
to be published
|
|
4LVK
 
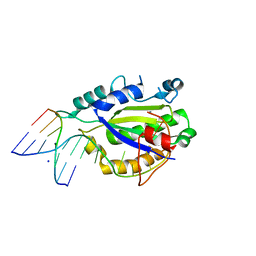 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Phosphate). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGpo oligonucleotide, MANGANESE (II) ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6C6T
 
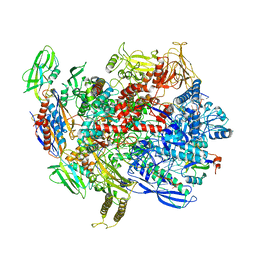 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6CAA
 
 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
6BPO
 
 | |
6BSQ
 
 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
3E29
 
 | | X-Ray structure of the protein Q7WE92_BORBR from thioesterase superfamily. Northeast Structural Genomics Consortium Target BoR214A. | | Descriptor: | uncharacterized protein Q7WE92_BORBR | | Authors: | Kuzin, A.P, Chen, Y, Setharaman, J, Wang, D, Mao, L, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-05 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray structure of the protein Q7WE92_BORBR from thioesterase superfamily. Northeast Structural Genomics Consortium Target BoR214A.
To be Published
|
|
6BVD
 
 | | Structure of Botulinum Neurotoxin Serotype HA Light Chain | | Descriptor: | ACETATE ION, CALCIUM ION, Light Chain, ... | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA.
Pathog Dis, 76, 2018
|
|
2Q2M
 
 | |
6DDQ
 
 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the basal state | | Descriptor: | 1,2-ETHANEDIOL, Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
