1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
6LLQ
 
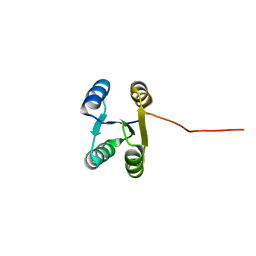 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | Descriptor: | VAL88 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MGZ
 
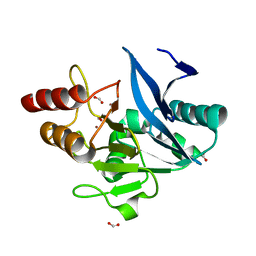 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NDM-4, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae
To Be Published
|
|
6MLJ
 
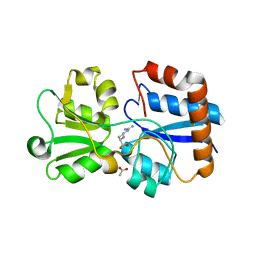 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) S70A mutant from Salmonella typhimurium complexed with arginine | | Descriptor: | ACETATE ION, ARGININE, Lysine/arginine/ornithine transport protein | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6MLV
 
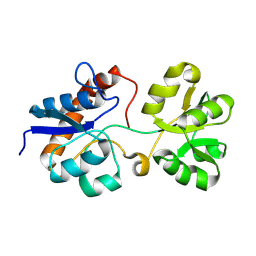 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) Y14A mutant from Salmonella typhimurium | | Descriptor: | Lysine/arginine/ornithine-binding periplasmic protein | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-28 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6MGX
 
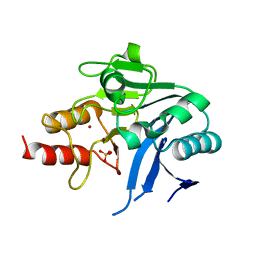 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae | | Descriptor: | Metallo-beta-lactamase, SULFATE ION, ZINC ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae
To Be Published
|
|
3RMF
 
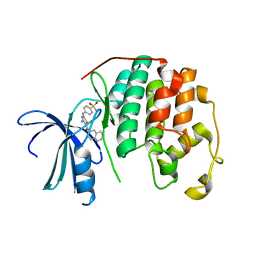 | | CDK2 in complex with inhibitor RC-2-33 | | Descriptor: | 4-{[4-amino-5-(naphthalen-2-ylcarbonyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
4IEL
 
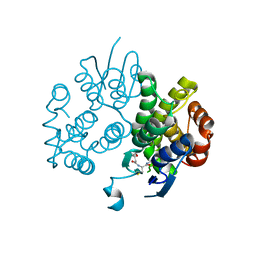 | | Crystal structure of a glutathione s-transferase family protein from burkholderia ambifaria, target efi-507141, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione s-transferase family protein from burkholderia ambifaria, target efi-507141, with bound glutathione
To be Published
|
|
6MGU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate
To Be Published
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4PQA
 
 | | Crystal Structure of succinyl-diaminopimelate desuccinylase from Neisseria meningitidis MC58 in complex with the Inhibitor Captopril | | Descriptor: | L-CAPTOPRIL, SULFATE ION, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Nocek, B, Starus, A, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
3R9N
 
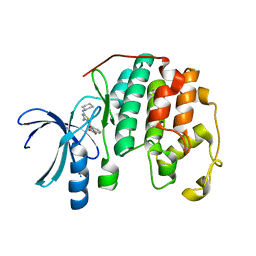 | | CDK2 in complex with inhibitor RC-2-21 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(cyclohexylamino)-1,3-thiazol-5-yl](3-nitrophenyl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
4CP4
 
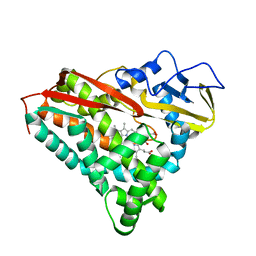 | |
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6ML0
 
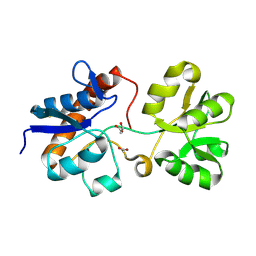 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) S69A mutant from Salmonella typhimurium | | Descriptor: | ACETATE ION, GLYCEROL, Lysine/arginine/ornithine-binding periplasmic protein | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6ML6
 
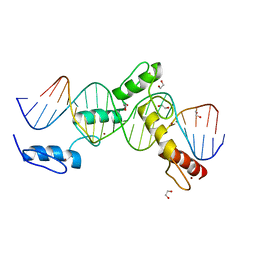 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
5AZU
 
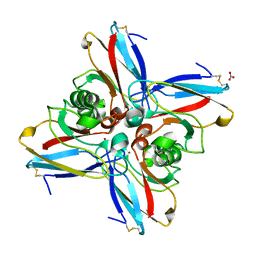 | |
6MLI
 
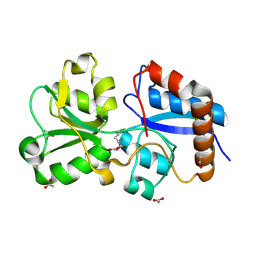 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) R77A mutant from Salmonella typhimurium complexed with histidine | | Descriptor: | ACETATE ION, GLYCEROL, HISTIDINE, ... | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6MLO
 
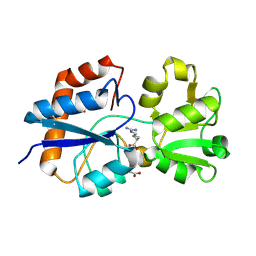 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) Y14A mutant from Salmonella typhimurium complexed with arginine | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6MLX
 
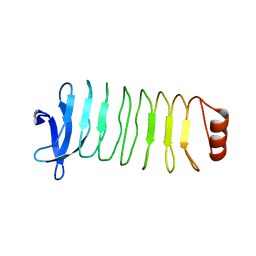 | | Crystal structure of T. pallidum Leucine Rich Repeat protein (TpLRR) | | Descriptor: | Leucine-rich repeat protein TpLRR | | Authors: | Ramaswamy, R, Loveless, B.C, Houston, S, Cameron, C.E, Boulanger, M.J. | | Deposit date: | 2018-09-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Treponema pallidum Tp0225 reveals an unexpected leucine-rich repeat architecture.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4PPI
 
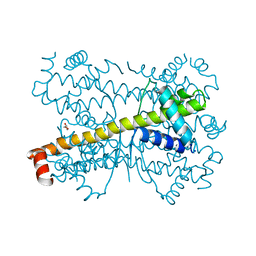 | | Crystal structure of Bcl-xL hexamer | | Descriptor: | Bcl-2-like protein 1, GLYCEROL | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
3RK0
 
 | | X-ray crystal Structure of the putative N-type ATP pyrophosphatase (PF0828) in complex with AMP from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR23 | | Descriptor: | ADENOSINE MONOPHOSPHATE, N-type ATP pyrophosphatase superfamily | | Authors: | Forouhar, F, Saadat, N, Hussain, M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Ma, L, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-16 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A large conformational change in the putative ATP pyrophosphatase PF0828 induced by ATP binding.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4CNI
 
 | | Crystal structure of the Fab portion of Olokizumab in complex with IL- 6 | | Descriptor: | INTERLEUKIN-6, OLOKIZUMAB HEAVY CHAIN, FAB PORTION, ... | | Authors: | Shaw, S, Bourne, T, Meier, C, Carrington, B, Gelinas, R, Henry, A, Popplewell, A, Adams, R, Baker, T, Rapecki, S, Marshall, D, Neale, H, Lawson, A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-30 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Characterization of Olokizumab: A Humanized Antibody Targeting Interleukin-6 and Neutralizing Gp130-Signaling.
Mabs, 6, 2014
|
|
3RK7
 
 | | CDK2 in complex with inhibitor RC-2-71 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(pyridin-3-ylcarbonyl)-1,3-thiazol-2-yl]amino}benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
6MGR
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine Monophosphate
To Be Published
|
|
