4M1E
 
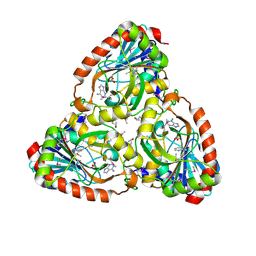 | | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364. | | Descriptor: | ADENINE, PYRIDINE-2-CARBOXYLIC ACID, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364.
To be Published
|
|
1E0V
 
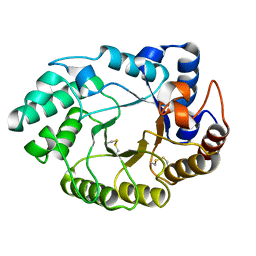 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
3CES
 
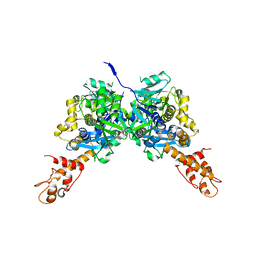 | | Crystal Structure of E.coli MnmG (GidA), a Highly-Conserved tRNA Modifying Enzyme | | Descriptor: | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
1MIT
 
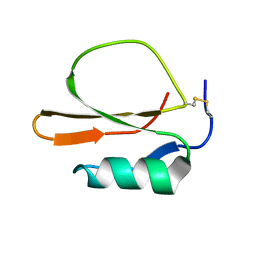 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
3USB
 
 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
5IB2
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IB5
 
 | | Crystal structure of HLA-B*27:09 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
1NJP
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
5AGC
 
 | | Crystallographic forms of the Vps75 tetramer | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 75 | | Authors: | Hammond, C.M, Sundaramoorthy, R, Owen-Hughes, T. | | Deposit date: | 2015-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Histone Chaperone Vps75 Forms Multiple Oligomeric Assemblies Capable of Mediating Exchange between Histone H3-H4 Tetramers and Asf1-H3-H4 Complexes.
Nucleic Acids Res., 44, 2016
|
|
1WXI
 
 | | E.coli NAD Synthetase, AMP.PP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
5LBB
 
 | |
5A4R
 
 | | Crystal structure of a vitamin B12 trafficking protein | | Descriptor: | METHYLMALONIC ACIDURIA AND HOMOCYSTINURIA TYPE D HOMOLOG, MITOCHONDRIAL | | Authors: | Kopec, J, Fitzpatrick, F, Froese, D.S, Velupillai, S, Nowak, R, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Fowler, B, Baumgartner, M.R, Yue, W.W. | | Deposit date: | 2015-06-11 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Mmachc-Mmadhc Protein Complex Involved in Vitamin B12 Trafficking.
J.Biol.Chem., 290, 2015
|
|
1NHG
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PLASMODIUM FALCIPARUM ENOYL-ACYL-CARRIER-PROTEIN REDUCTASE WITH TRICLOSAN | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN, enoyl-acyl carrier reductase | | Authors: | Perozzo, R, Kuo, M, Sidhu, A.S, Valiyaveettil, J.T, Bittman, R, Jacobs Jr, W.R, Fidock, D.A, Sacchettini, J.C. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Elucidation of the Specificity of the Antibacterial Agent Triclosan for
Malarial Enoyl Acyl Carrier Protein Reductase
J.Biol.Chem., 277, 2002
|
|
1WLH
 
 | | Molecular structure of the rod domain of Dictyostelium filamin | | Descriptor: | Gelation factor | | Authors: | Popowicz, G.M, Mueller, R, Noegel, A.A, Schleicher, M, Huber, R, Holak, T.A. | | Deposit date: | 2004-06-27 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of the rod domain of dictyostelium filamin
J.Mol.Biol., 342, 2004
|
|
1NJN
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with the antibiotic sparsomycin | | Descriptor: | 23S ribosomal RNA, SPARSOMYCIN | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
5L9B
 
 | |
5LA9
 
 | |
5L9R
 
 | |
5LBF
 
 | |
5LAT
 
 | |
1WXG
 
 | | E.coli NAD Synthetase, DND | | Descriptor: | MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
6UTL
 
 | | Yeast Thiol Specific antoxidant 2 with C171S mutation and catalytic cysteine alkylated with iodoacetamide | | Descriptor: | Peroxiredoxin TSA2 | | Authors: | Tairum, C.A, Bannitz-Fernandes, R, Tonoli, C.C.C, Murakami, M.T, de Oliveira, M.A, Netto, L.E.S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduction of sulfenic acids by ascorbate in proteins, connecting thiol-dependent to alternative redox pathways.
Free Radic Biol Med, 156, 2020
|
|
1WWR
 
 | | Crystal structure of tRNA adenosine deaminase TadA from Aquifex aeolicus | | Descriptor: | ZINC ION, tRNA adenosine deaminase TadA | | Authors: | Kuratani, M, Ishii, R, Bessho, Y, Fukunaga, R, Sengoku, T, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of tRNA Adenosine Deaminase (TadA) from Aquifex aeolicus
J.Biol.Chem., 280, 2005
|
|
1IEG
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT S134A/H157A OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
