1XW8
 
 | | X-ray structure of putative lactam utilization protein YBGL. Northeast Structural Genomics Consortium target ET90. | | Descriptor: | UPF0271 protein ybgL | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S.M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of putative lactam utilization protein YBGL.
Northeast Structural Genomics Consortium target ET90.
To be Published
|
|
4KTE
 
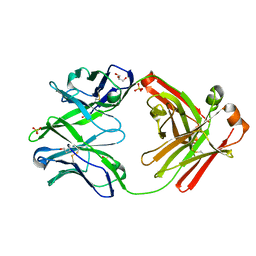 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6GC6
 
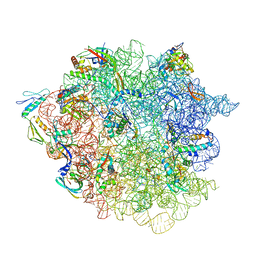 | | 50S ribosomal subunit assembly intermediate state 2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
1XVL
 
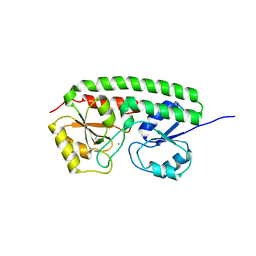 | | The three-dimensional structure of MntC from Synechocystis 6803 | | Descriptor: | MANGANESE (II) ION, Mn transporter | | Authors: | Rukhman, V, Anati, R, Melamed-Frank, M, Bhattacharyya-Pakrasi, M, Pakrasi, H.B, Adir, N. | | Deposit date: | 2004-10-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The MntC crystal structure suggests that import of Mn2+ in cyanobacteria is redox controlled.
J.Mol.Biol., 348, 2005
|
|
2AN1
 
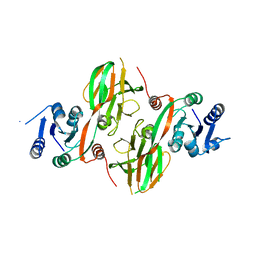 | | Structural Genomics, The crystal structure of a putative kinase from Salmonella typhimurim LT2 | | Descriptor: | putative kinase | | Authors: | Zhang, R, Zhou, M, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a putative kinase from Salmonella typhimurim LT2
To be Published
|
|
1KLA
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1XXN
 
 | | Crystal structure of a mesophilic xylanase A from Bacillus subtilis 1A1 | | Descriptor: | Endo-1,4-beta-xylanase A, S,R MESO-TARTARIC ACID | | Authors: | Murakami, M.T, Ruller, R, Ward, R.J, Arni, R.K. | | Deposit date: | 2004-11-07 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Correlation of temperature induced conformation change with optimum catalytic activity in the recombinant G/11 xylanase A from Bacillus subtilis strain 168 (1A1).
Febs Lett., 579, 2005
|
|
4AOC
 
 | | crystal structure of BC2L-A Lectin from Burkolderia cenocepacia in complex with methyl-heptoside | | Descriptor: | BC2L-A LECTIN, CALCIUM ION, SULFATE ION, ... | | Authors: | Marchetti, R, Malinovska, L, Lameignere, E, deCastro, C, Cioci, G, Kosma, P, Wimmerova, M, Molinaro, A, Imberty, A, Silipo, A. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Burkholderia Cenocepacia Lectin a Binding to Heptoses from the Bacterial Lipopolysaccharide.
Glycobiology, 22, 2012
|
|
1XY9
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
4APP
 
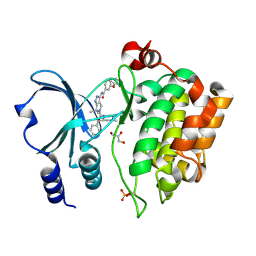 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with (S)-N-(5-(3-benzyl-1-methylpiperazine-4-carbonyl)-6,6-dimethyl-1,4,5, 6-tetrahydropyrrolo(3,4-c)pyrazol-3-yl)-3-phenoxybenzamide | | Descriptor: | GLYCEROL, N-[6,6-dimethyl-5-[(2S)-4-methyl-2-(phenylmethyl)piperazin-1-yl]carbonyl-2,4-dihydropyrrolo[3,4-c]pyrazol-3-yl]-3-phenoxy-benzamide, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.D, Deng, Y.L, Wang, C, Guo, C, McAlpine, I, Zhang, J, Kephart, S, Johnson, M.C, Li, H, Bouzida, D, Yang, A, Dong, L, Marakovits, J, Tikhe, J, Richardson, P, Guo, L.C, Kania, R, Edwards, M.P, Kraynov, E, Christensen, J, Piraino, J, Lee, J, Dagostino, E, Del-Carmen, C, Smeal, T, Murray, B.W. | | Deposit date: | 2012-04-04 | | Release date: | 2012-06-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Pyrroloaminopyrazoles as Novel Pak Inhibitors.
J.Med.Chem., 55, 2012
|
|
2APW
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
1XZ3
 
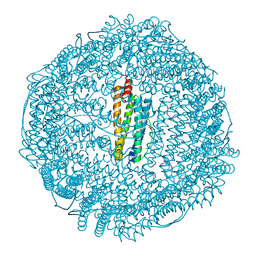 | | Complex of apoferritin with isoflurane | | Descriptor: | 1-CHLORO-2,2,2-TRIFLUOROETHYL DIFLUOROMETHYL ETHER, CADMIUM ION, Ferritin light chain | | Authors: | Liu, R, Loll, P.J, Eckenhoff, R.G. | | Deposit date: | 2004-11-11 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein.
Faseb J., 19, 2005
|
|
4TOB
 
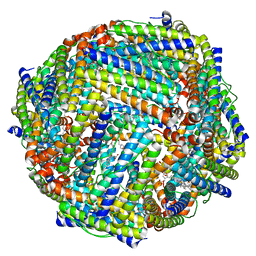 | | 1.95A resolution structure of BfrB (Q151L) from Pseudomonas aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bacterioferritin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4BCC
 
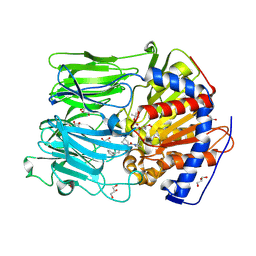 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND P2- substituted N-acyl-prolylpyrrolidine inhibitor | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | VanDerVeken, P, Fulop, V, Rea, D, Gerard, M, VanElzen, R, Joossens, J, Cheng, J.D, Baekelandt, V, DeMeester, I, Lambeir, A.M, Augustyns, K. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | P2-Substituted N-Acylprolylpyrrolidine Inhibitors of Prolyl Oligopeptidase: Biochemical Evaluation, Binding Mode Determination, and Assessment in a Cellular Model of Synucleinopathy.
J.Med.Chem., 55, 2012
|
|
1KP7
 
 | | Conserved RNA Structure within the HCV IRES eIF3 Binding Site | | Descriptor: | Hepatitis C Virus Internal Ribosome Entry Site Fragment | | Authors: | Gallego, J, Klinck, R, Collier, A.J, Cole, P.T, Harris, S.J, Harrison, G.P, Aboul-ela, F, Walker, S, Varani, G. | | Deposit date: | 2001-12-29 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conserved RNA structure within the HCV IRES eIF3-binding site.
Nat.Struct.Biol., 9, 2002
|
|
4TR5
 
 | |
1XJJ
 
 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dGTP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ribonucleotide reductase, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Mechanism of Allosteric Substrate Specificity Regulation in a Ribonucleotide Reductase
Nat.Struct.Mol.Biol., 11, 2005
|
|
4RWS
 
 | | Crystal structure of CXCR4 and viral chemokine antagonist vMIP-II complex (PSI Community Target) | | Descriptor: | C-X-C chemokine receptor type 4/Endolysin chimeric protein, Viral macrophage inflammatory protein 2 | | Authors: | Qin, L, Kufareva, I, Holden, L, Wang, C, Zheng, Y, Wu, H, Fenalti, G, Han, G.W, Cherezov, V, Abagyan, R, Stevens, R.C, Handel, T.M, GPCR Network (GPCR) | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Science, 347, 2015
|
|
2B7D
 
 | | Factor VIIa Inhibitors: Chemical Optimization, Preclinical Pharmacokinetics, Pharmacodynamics, and Efficacy in a Baboon Thrombosis Model | | Descriptor: | (2R)-2-[5-(5-CARBAMIMIDOYL-1H-BENZOIMIDAZOL-2-YL)-6,2'-DIHYDROXY-5'-UREIDOMETHYL-BIPHENYL-3-YL]-SUCCINIC ACID, Coagulation factor VII, Tissue factor | | Authors: | Young, W.B, Mordenti, J, Torkelson, S, Shrader, W.D, Kolesnikov, A, Rai, R, Liu, L, Hu, H, Leahy, E.M, Green, M.J, Sprengeler, P.A, Katz, B.A, Yu, C, Janc, J.W, Elrod, K.C, Marzec, U.M, Hanson, S.R. | | Deposit date: | 2005-10-04 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Factor VIIa inhibitors: Chemical optimization, preclinical pharmacokinetics, pharmacodynamics, and efficacy in an arterial baboon thrombosis model.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1XM3
 
 | | Crystal structure of Northeast Structural Genomics Target SR156 | | Descriptor: | Thiazole biosynthesis protein thiG | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S, Forouhar, F, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Northeast Structural Genomics Target SR156
To be Published
|
|
1XJG
 
 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dATP-UDP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism of Allosteric Substrate Specificity Regulation in a Ribonucleotide Reductase
Nat.Struct.Mol.Biol., 11, 2004
|
|
4RZ0
 
 | | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c | | Descriptor: | PFL0690c, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-17 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c
To be Published
|
|
4MX6
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella oneidensis (SO_3134), target EFI-510275, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP-type C4-dicarboxylate:H+ symport system substrate-binding component DctP | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-25 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1XN8
 
 | | Solution Structure of Bacillus subtilis Protein yqbG: The Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Ma, L, Shen, Y, Acton, T, Atreya, H.S, Xiao, R, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6DZN
 
 | | Pan-ebolavirus human antibody ADI-15878 Fab | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, Antibody ADI-15878, heavy chain, ... | | Authors: | Murin, C.D, Ward, A.B, Bruhn, J.F, Stanfield, R. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Pan-Ebolavirus Neutralization by an Antibody Targeting the Glycoprotein Fusion Loop.
Cell Rep, 24, 2018
|
|
