7Q56
 
 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase (minor conformer) from Spinacia oleracea. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q3M
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 3-hydroxypiperidine | | Descriptor: | (3S)-piperidin-3-ol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7Q2V
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1-methylpyrrolidin-3-ol | | Descriptor: | (3R)-1-methylpyrrolidin-3-ol, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7SNI
 
 | | Structure of G6PD-D200N tetramer bound to NADP+ and G6P | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q2K
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 2-pyrrolidinone | | Descriptor: | Cholinephosphate cytidylyltransferase, pyrrolidin-2-one | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7Q3W
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with (R)-2-Aminobutanamide hydrochloride | | Descriptor: | (R)-2-Aminobutanamide, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7SNG
 
 | | structure of G6PD-WT tetramer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6FF7
 
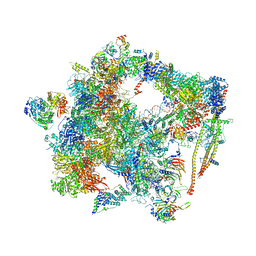 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
3WNL
 
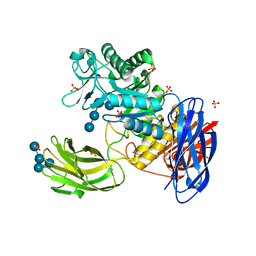 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
7SX5
 
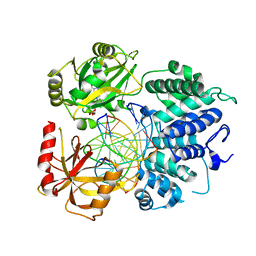 | | Crystal structure of ligase I with nick duplexes containing mismatch A:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA chain 1, DNA chain 2, ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
7Q57
 
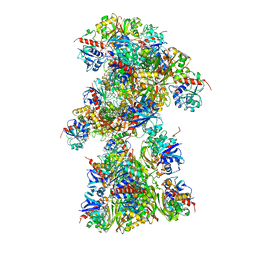 | | Single Particle Cryo-EM structure of photosynthetic A10B10 glyceraldehyde-3-phospahte dehydrogenase from Spinacia oleracea. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8RUU
 
 | | Fabs derived from bimekizumab in complex with IL-17F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoblobulin heavy chain, Immunoblobulin light chain, ... | | Authors: | Adams, R, Lawson, A.D.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Bimekizumab Fab Fragment in Complex with IL-17F Provides Molecular Basis for Dual IL-17A and IL-17F Inhibition.
J Invest Dermatol., 2024
|
|
4XMW
 
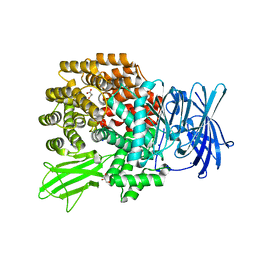 | |
4XVO
 
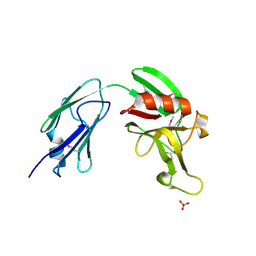 | | L,D-transpeptidase from Mycobacterium smegmatis | | Descriptor: | L,D-transpeptidase, PHOSPHATE ION | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-27 | | Release date: | 2015-02-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L,D-transpeptidase from Mycobacterium smegmatis
to be published
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PQU
 
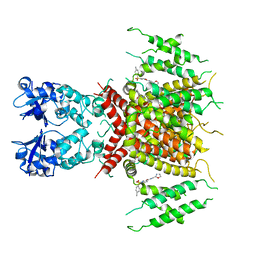 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | Descriptor: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7SXE
 
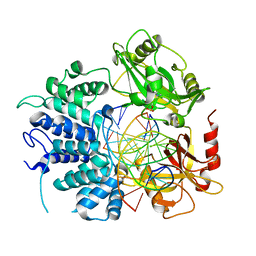 | | Crystal structure of ligase I with nick duplexes containing cognate G:T | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA chain 1, DNA chain 2, ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
8SJV
 
 | | [4T24] Self-assembling left-handed tensegrity triangle with 24 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*TP*TP*GP*TP*AP*GP*TP*CP*TP*CP*AP*CP*CP*AP*CP*TP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*CP*AP*CP*TP*CP*CP*TP*GP*AP*GP*AP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*CP*AP*TP*CP*AP*CP*AP*GP*TP*GP*GP*AP*CP*TP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.59 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4XWY
 
 | | Crystal structure of human sepiapterin reductase in complex with an N-acetylserotinin analogue | | Descriptor: | N-[2-(5-hydroxy-2-methyl-1H-indol-3-yl)ethyl]-2-methoxyacetamide, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Johnsson, K, Hovius, R, Gorszka, K.I, Pojer, F. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Reduction of Neuropathic and Inflammatory Pain through Inhibition of the Tetrahydrobiopterin Pathway.
Neuron, 86, 2015
|
|
7Q2I
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Tetrahydrofurfurylamine | | Descriptor: | 1-[(2R)-oxolan-2-yl]methanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q53
 
 | | Single Particle Cryo-EM structure of photosynthetic A2B2 glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q5V
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH N-OXALYLGLYCINE (NOG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
7Q2L
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 3-Aminopyrrolidin-2-one hydrochloride | | Descriptor: | (3R)-3-aminopyrrolidin-2-one, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
6PJK
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-29 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-isoleucyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
7Q2M
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with but-3-yn-2-amine hydrochloride | | Descriptor: | (2S)-but-3-yn-2-amine, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
