3OSJ
 
 | | X-Ray Structure of Phycobilisome LCM core-membrane linker polypeptide (fragment 254-400) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR209C | | Descriptor: | NITRATE ION, Phycobilisome LCM core-membrane linker polypeptide, SODIUM ION | | Authors: | Kuzin, A, Su, M, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-09 | | Release date: | 2010-10-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR209C
To be published
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3LMC
 
 | | Crystal Structure of zinc-dependent peptidase from Methanocorpusculum labreanum (strain Z), Northeast Structural Genomics Consortium Target MuR16 | | Descriptor: | FE (III) ION, Peptidase, zinc-dependent, ... | | Authors: | Kuzin, A, Ashok, S, Vorobiev, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Northeast Structural Genomics Consortium Target MuR16
To be published
|
|
3LML
 
 | | Crystal Structure of the sheath tail protein Lin1278 from Listeria innocua, Northeast Structural Genomics Consortium Target LkR115 | | Descriptor: | Lin1278 protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target LkR115
To be Published
|
|
4J7Z
 
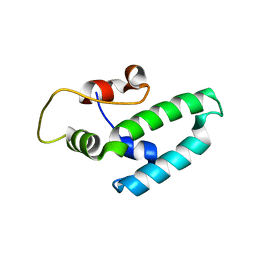 | | Thermus thermophilus DNAJ J- and G/F-DOMAINS | | Descriptor: | Chaperone protein DnaJ 2, GLYCEROL | | Authors: | Barends, T.R.M, Brosi, R.W, Steinmetz, A, Scherer, A, Hartmann, E, Eschenbach, J, Lorenz, T, Seidel, R, Shoeman, R, Zimmermann, S, Bittl, R, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | EPR (1.64 Å), X-RAY DIFFRACTION | | Cite: | Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5FXQ
 
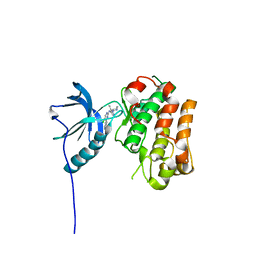 | | IGFR-1R complex with a pyrimidine inhibitor. | | Descriptor: | 5-chloranyl-4-imidazo[1,2-a]pyridin-3-yl-N-(5-methyl-1-piperidin-4-yl-pyrazol-4-yl)pyrimidin-2-amine, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR | | Authors: | Degorce, S, Boyd, S, Curwen, J, Ducray, R, Halsall, C, Jones, C, Lach, F, Lenz, E, Pass, M, Pass, S, Trigwell, C, Norman, R, Phillips, C. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Potent, Selective, Orally Bioavailable, and Efficacious Novel 2-(Pyrazol-4-ylamino)-pyrimidine Inhibitor of the Insulin-like Growth Factor-1 Receptor (IGF-1R).
J. Med. Chem., 59, 2016
|
|
1PZX
 
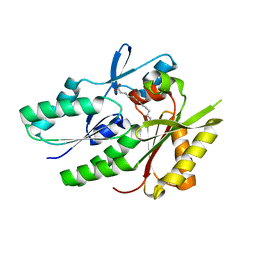 | | Hypothetical protein APC36103 from Bacillus stearothermophilus: a lipid binding protein | | Descriptor: | Hypothetical protein APC36103, PALMITIC ACID | | Authors: | Zhang, R, Osipiuk, J, Zhou, M, Alkire, R, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-07-14 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid binding protein APC36103 from Bacillus Stearothermophilus
To be Published
|
|
4Q4E
 
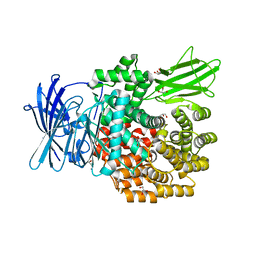 | | Crystal structure of E.coli aminopeptidase N in complex with actinonin | | Descriptor: | ACTINONIN, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
2WWC
 
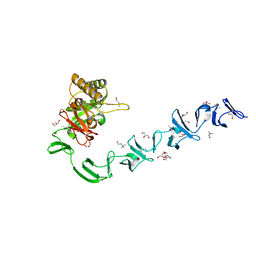 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with synthetic peptidoglycan ligand | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHOLINE ION, GLYCEROL | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4C01
 
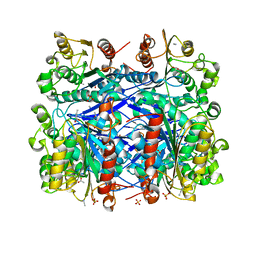 | | Complete crystal structure of carboxylesterase Cest-2923 (lp_2923) from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, ACETONITRILE, CEST-2923, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
4N8Z
 
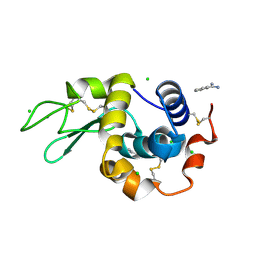 | | In situ lysozyme crystallized on a MiTeGen micromesh with benzamidine ligand | | Descriptor: | BENZAMIDINE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ co-crystallization of proteins plus fragment libraries on pin-mounted data-collection micromeshes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MPF
 
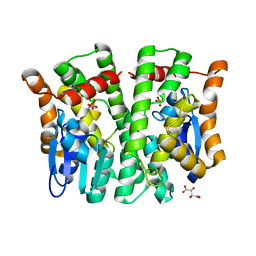 | | Crystal structure of human glutathione transferase theta-2, complex with inorganic phosphate, GSH free, target EFI-507257 | | Descriptor: | Glutathione S-transferase theta-2, L(+)-TARTARIC ACID, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Glutathione S-Transferase Theta-2 (Target Efi-507257)
To be Published
|
|
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
3KKY
 
 | | Structure of Manganese Superoxide Dismutase from Deinococcus Radiodurans in the orthorhombic space group P212121: A case study of mistaken identity | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Govindasamy, L, Mikulski, R, McKenna, M.A, Silverman, D.N, McKenna, R. | | Deposit date: | 2009-11-06 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Manganese Superoxide dismutase from Deinococcus Radiodurans in the orthorhombic space group P212121: A case study of mistaken identity
To be Published
|
|
4F3S
 
 | | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica | | Descriptor: | D-ALANINE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Foti, R, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica
To be Published
|
|
4OG1
 
 | | Crystal Structure of a Putative Enoyl-CoA Hydratase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Enoyl-CoA hydratase/isomerase | | Authors: | Chapman, H.C, Cooper, D.R, Tkaczuk, K.L, Cymborowski, M.T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Alkire, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of a Putative Enoyl-CoA Hydratase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4OOA
 
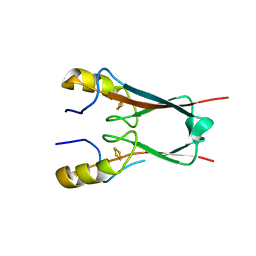 | | CRYSTAL STRUCTURE of NAF1 (MINER1): H114C THE REDOX-ACTIVE 2FE-2S PROTEIN | | Descriptor: | CDGSH iron-sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Tamir, S, Eisenberg-Domovich, Y, Conlan, A.R, Stofleth, J.T, Lipper, C.H, Paddock, M.L, Mittler, R, Jennings, P.A, Livnah, O, Nechushtai, R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AQX
 
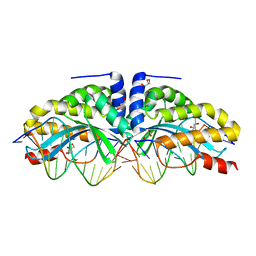 | | Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of magnesium | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*5CM)-3', 5'-D(*GP*AP*CP*AP*GP*TP*TP*TP*GP*GP)-3', 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', ... | | Authors: | Valton, J, Daboussi, F, Leduc, S, Redondo, P, Macmaster, R, Molina, R, Montoya, G, Duchateau, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases.
J.Biol.Chem., 287, 2012
|
|
1P8J
 
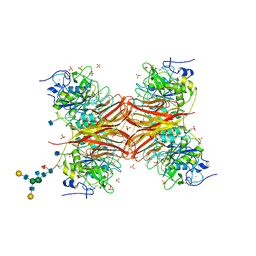 | | CRYSTAL STRUCTURE OF THE PROPROTEIN CONVERTASE FURIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Henrich, S, Cameron, A, Bourenkov, G.P, Kiefersauer, R, Huber, R, Lindberg, I, Bode, W, Than, M.E. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Proprotein Processing Proteinase Furin Explains its Stringent Specificity
Nat.Struct.Biol., 10, 2003
|
|
3NIX
 
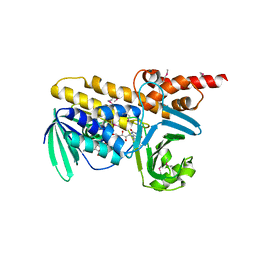 | | Crystal structure of flavoprotein/dehydrogenase from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target ChR43. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavoprotein/dehydrogenase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of flavoprotein/dehydrogenase from Cytophaga hutchinsonii.
To be Published
|
|
3NKY
 
 | | Structure of a mutant P44S of Foot-and-mouth disease Virus RNA-dependent RNA polymerase | | Descriptor: | 3D polymerase, MAGNESIUM ION | | Authors: | Agudo, R, Ferrer-Orta, C, Arias, A, Perez-Luque, R, Verdaguer, N, Domingo, E. | | Deposit date: | 2010-06-21 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape.
Plos Pathog., 6, 2010
|
|
4FBY
 
 | | fs X-ray diffraction of Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Hellmich, J, Tran, R, Hattne, J, Laksmono, H, Gloeckner, C, Echols, N, Sierra, R.G, Sellberg, J, Lassalle-Kaiser, B, Gildea, R.J, Glatzel, P, Grosse-Kunstleve, R.W, Latimer, M.J, Mcqueen, T.A, Difiore, D, Fry, A.R, Messerschmidt, M.M, Miahnahri, A, Schafer, D.W, Seibert, M.M, Sokaras, D, Weng, T.-C, Zwart, P.H, White, W.E, Adams, P.D, Bogan, M.J, Boutet, S, Williams, G.J, Messinger, J, Sauter, N.K, Zouni, A, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (6.56 Å) | | Cite: | Room temperature femtosecond X-ray diffraction of photosystem II microcrystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1OTT
 
 | | Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
4DYK
 
 | | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn
to be published
|
|
