3H5I
 
 | | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans | | Descriptor: | CHLORIDE ION, Response regulator/sensory box protein/GGDEF domain protein, SODIUM ION | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans
To be Published
|
|
4Z9Y
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Pectobacterium carotovorum | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, SULFATE ION | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
2L2S
 
 | |
2L3H
 
 | | NMR Structure in a Membrane Environment Reveals Putative Amyloidogenic Regions of the SEVI Precursor Peptide PAP248-286 | | Descriptor: | Prostatic acid phosphatase | | Authors: | Ramamoorthy, A, Nanga, R, Brender, J, Vivekanandan, S, Popovych, N. | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure in a membrane environment reveals putative amyloidogenic regions of the SEVI precursor peptide PAP(248-286).
J.Am.Chem.Soc., 131, 2009
|
|
2L7V
 
 | | Quindoline/G-quadruplex complex | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), N,N-diethyl-N'-(10H-indolo[3,2-b]quinolin-11-yl)ethane-1,2-diamine, POTASSIUM ION | | Authors: | Dai, J, Carver, M, Mathad, R, Yang, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a 2:1 Quindoline-c-MYC G-Quadruplex: Insights into G-Quadruplex-Interactive Small Molecule Drug Design.
J.Am.Chem.Soc., 133, 2011
|
|
4ZEO
 
 | | Crystal structure of eIF2B delta from Chaetomium thermophilum | | Descriptor: | Translation initiation factor eif-2b-like protein | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2.
Nucleic Acids Res., 43, 2015
|
|
4TW1
 
 | | Crystal structure of the octameric pore complex of the Staphylococcus aureus Bi-component Toxin LukGH | | Descriptor: | Possible leukocidin subunit | | Authors: | Logan, D.T, Hakansson, M, Saline, M, Kimbung, R, Badarau, A, Rouha, H, Nagy, E. | | Deposit date: | 2014-06-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Function Analysis of Heterodimer Formation, Oligomerization, and Receptor Binding of the Staphylococcus aureus Bi-component Toxin LukGH.
J.Biol.Chem., 290, 2015
|
|
2LNV
 
 | |
6ICE
 
 | | Crystal structure of Hamster MIF | | Descriptor: | Macrophage migration inhibitory factor | | Authors: | Sundaram, R, Vasudevan, D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
4ZI9
 
 | | Structure of mouse clustered PcdhgA1 EC1-3 | | Descriptor: | CALCIUM ION, MCG133388, isoform CRA_t | | Authors: | Nicoludis, J.M, Lau, S.-Y, Scharfe, C.P.I, Marks, D.S, Weihofen, W.A, Gaudet, R. | | Deposit date: | 2015-04-27 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Structure, 23, 2015
|
|
4ZIX
 
 | | Structure of HEWL using Serial Femtosecond Crystallography of Soluble Proteins in Lipidic Cubic Phase | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Fromme, R, Ishchenko, A, Metz, M, Roy-Chowdhury, S, Basu, S, Boutet, S, Fromme, P, White, T.A, Barty, A, Spence, J.C.H, Weierstall, U, Liu, W, Cherezov, V. | | Deposit date: | 2015-04-28 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Serial Femtosecond Crystallography of Soluble Proteins in Lipidic Cubic Phase
IUCrJ, 2, 2015
|
|
2L86
 
 | |
4ZO6
 
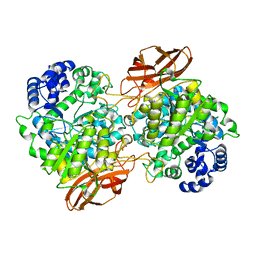 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with cellobiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
2LHZ
 
 | | Di-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
4ZO7
 
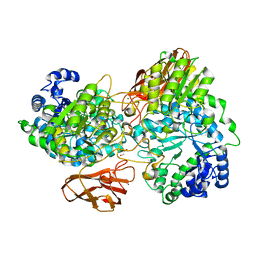 | | Crystal structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with gentiobiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
6IOL
 
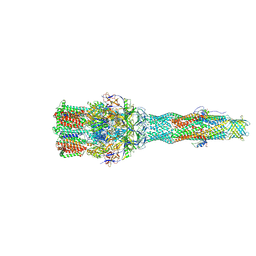 | | Cryo-EM structure of multidrug efflux pump MexAB-OprM (60 degree state) | | Descriptor: | Multidrug resistance protein MexA, Multidrug resistance protein MexB, Outer membrane protein OprM | | Authors: | Tsutsumi, K, Yonehara, R, Nakagawa, A, Yamashita, E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism.
Nat Commun, 10, 2019
|
|
6IDF
 
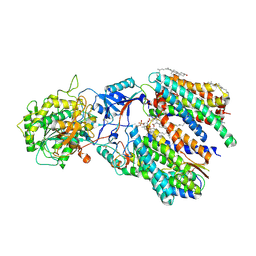 | | Cryo-EM structure of gamma secretase in complex with a Notch fragment | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Zhou, Q, Guo, X, Yan, C, Ke, M, Lei, J, Shi, Y. | | Deposit date: | 2018-09-09 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of Notch recognition by human gamma-secretase
Nature, 565, 2019
|
|
6IRV
 
 | |
6IF2
 
 | | Complex structure of Rab35 and its effector RUSC2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Iporin, MAGNESIUM ION, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6IFG
 
 | | Crystal structure of M1 zinc metallopeptidase E323A mutant bound to Tyr-ser-ala substrate from Deinococcus radiodurans | | Descriptor: | FORMIC ACID, Tripeptides (TYR-SER-ALA), ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two-domain aminopeptidase of M1 family: Structural features for substrate binding and gating in absence of C-terminal domain.
J.Struct.Biol., 208, 2019
|
|
6IUY
 
 | | Structure of DsGPDH of Dunaliella salina | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Glycerol-3-phosphate dehydrogenase [NAD(+)], ... | | Authors: | He, Q, Toh, J.D, Ero, R, Qiao, Z, Kumar, V, Gao, Y.G. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The unusual di-domain structure of Dunaliella salina glycerol-3-phosphate dehydrogenase enables direct conversion of dihydroxyacetone phosphate to glycerol.
Plant J., 102, 2020
|
|
4ZM0
 
 | |
2L3S
 
 | | Structure of the autoinhibited Crk | | Descriptor: | Autoinhibited Crk protein | | Authors: | Kalodimos, C.G, Sarkar, P, Saleh, T, Tzeng, S.R, Birge, R. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for regulation of the Crk signaling protein by a proline switch.
Nat.Chem.Biol., 7, 2011
|
|
4ZA2
 
 | | Crystal structure of Pectobacterium carotovorum 2-keto-3-deoxy-D-gluconate dehydrogenase complexed with NAD+ | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4ZNW
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 4-Bromo-substituted OBHS derivative | | Descriptor: | 4-bromophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
