3ZZ3
 
 | | Crystal structure of 3C protease mutant (N126Y) of coxsackievirus B3 | | 分子名称: | 3C PROTEINASE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-31 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
6L12
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
1TGS
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX BETWEEN PANCREATIC SECRETORY INHIBITOR (KAZAL TYPE) AND TRYPSINOGEN AT 1.8 ANGSTROMS RESOLUTION. STRUCTURE SOLUTION, CRYSTALLOGRAPHIC REFINEMENT AND PRELIMINARY STRUCTURAL INTERPRETATION | | 分子名称: | CALCIUM ION, PANCREATIC SECRETORY TRYPSIN INHIBITOR (KAZAL TYPE), SULFATE ION, ... | | 著者 | Bolognesi, M, Gatti, G, Menegatti, E, Guarneri, M, Marquart, M, Papamokos, E, Huber, R. | | 登録日 | 1982-09-27 | | 公開日 | 1983-01-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Three-dimensional structure of the complex between pancreatic secretory trypsin inhibitor (Kazal type) and trypsinogen at 1.8 A resolution. Structure solution, crystallographic refinement and preliminary structural interpretation.
J.Mol.Biol., 162, 1982
|
|
3ZD0
 
 | | The Solution Structure of Monomeric Hepatitis C Virus p7 Yields Potent Inhibitors of Virion Release | | 分子名称: | P7 PROTEIN | | 著者 | Foster, T.L, Sthompson, G, Kalverda, A.P, Kankanala, J, Thompson, J, Barker, A.M, Clarke, D, Noerenberg, M, Pearson, A.R, Rowlands, D.J, Homans, S.W, Harris, M, Foster, R, Griffin, S.D.C. | | 登録日 | 2012-11-23 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Guided Design Affirms Inhibitors of Hepatitis C Virus P7 as a Viable Class of Antivirals Targeting Virion Release
Hepatology, 59, 2014
|
|
6KYE
 
 | | The crystal structure of recombinant human adult hemoglobin | | 分子名称: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | 著者 | Kihira, K, Funaki, R, Okamoto, W, Endo, C, Morita, Y, Komatsu, T. | | 登録日 | 2019-09-18 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Genetically engineered haemoglobin wrapped covalently with human serum albumins as an artificial O2carrier.
J Mater Chem B, 8, 2020
|
|
6HYP
 
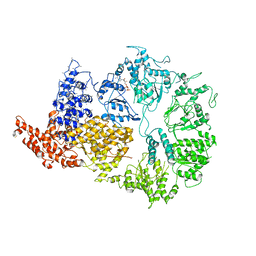 | | Rea1 Wild type ADP state (AAA+ ring part) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Midasin,Midasin | | 著者 | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | 登録日 | 2018-10-22 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
4FP7
 
 | |
1TU5
 
 | | Crystal structure of bovine plasma copper-containing amine oxidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Lunelli, M, Di Paolo, M.L, Biadene, M, Calderone, V, Scarpa, M, Battistutta, R, Rigo, A, Zanotti, G. | | 登録日 | 2004-06-24 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Crystal structure of amine oxidase from bovine serum.
J.Mol.Biol., 346, 2005
|
|
3ZCR
 
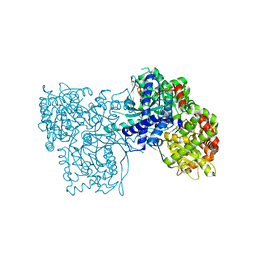 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4-tert- butyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | 分子名称: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | 著者 | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | 登録日 | 2012-11-21 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZFG
 
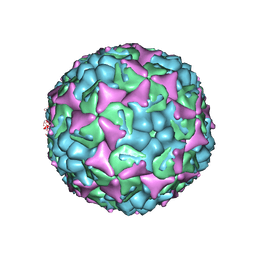 | | Human enterovirus 71 in complex with capsid binding inhibitor WIN51711 | | 分子名称: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, VP1, VP2, ... | | 著者 | Plevka, P, Perera, R, Yap, M.L, Cardosa, J, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2012-12-11 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of Human Enterovirus 71 in Complex with a Capsid-Binding Inhibitor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZEI
 
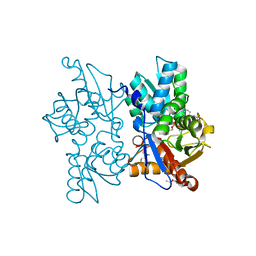 | | Structure of the Mycobacterium tuberculosis O-Acetylserine Sulfhydrylase (OASS) CysK1 in complex with a small molecule inhibitor | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(Z)-[(5Z)-5-[[2-(2-hydroxy-2-oxoethyloxy)phenyl]methylidene]-3-methyl-4-oxidanylidene-1,3-thiazolidin-2-ylidene]amino]benzoic acid, O-ACETYLSERINE SULFHYDRYLASE, ... | | 著者 | Poyraz, O, Schnell, R, Schneider, G. | | 登録日 | 2012-12-05 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Guided Design of Novel Thiazolidine Inhibitors of O-Acetyl Serine Sulfhydrylase from Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
3ZFC
 
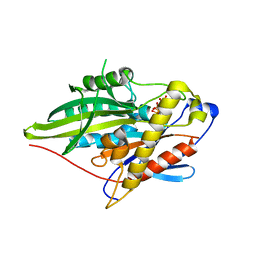 | | Crystal Structure of the Kif4 Motor Domain Complexed With Mg-AMPPNP | | 分子名称: | CHROMOSOME-ASSOCIATED KINESIN KIF4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Chang, Q, Nitta, R, Inoue, S, Hirokawa, N. | | 登録日 | 2012-12-11 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for the ATP-Induced Isomerization of Kinesin.
J.Mol.Biol., 425, 2013
|
|
3ZC3
 
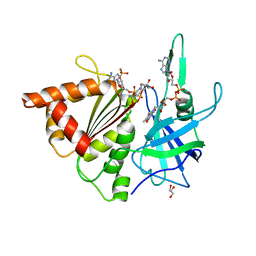 | | FERREDOXIN-NADP REDUCTASE (MUTATION S80A) COMPLEXED WITH NADP BY COCRYSTALLIZATION | | 分子名称: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Martinez-Julvez, M, Hurtado-Guerrero, R, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | 登録日 | 2012-11-15 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
4F93
 
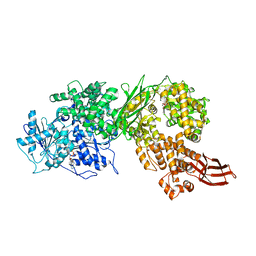 | | Brr2 Helicase Region S1087L, Mg-ATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | 登録日 | 2012-05-18 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | 分子名称: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | 登録日 | 2020-08-19 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3ZCQ
 
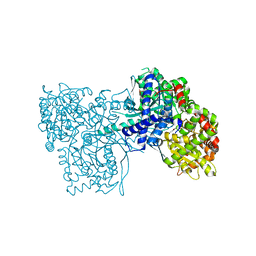 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4- trifluoromethyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2. 15 A resolution | | 分子名称: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-{[4-(trifluoromethyl)benzoyl]carbamoyl}-beta-D-glucopyranosylamine, ... | | 著者 | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | 登録日 | 2012-11-21 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
7JO3
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)pivalamide | | 分子名称: | 2,2-dimethyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Andring, J.T, McKenna, R. | | 登録日 | 2020-08-05 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.454 Å) | | 主引用文献 | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
4N1C
 
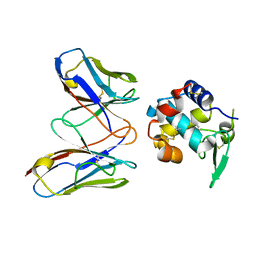 | | Structural evidence for antigen receptor evolution | | 分子名称: | Lysozyme C, immunoglobulin variable light chain domain | | 著者 | Langley, D.B, Rouet, R, Roome, B, Stock, D, Christ, D. | | 登録日 | 2013-10-03 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6I1T
 
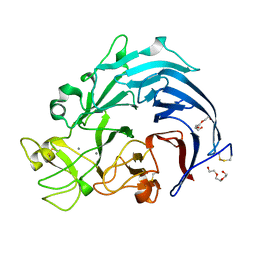 | | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Turbe-Doan, A, Record, E, Lombard, V, Kumar, R, Henrissat, B, Levasseur, A, Garron, M.L. | | 登録日 | 2018-10-30 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The functional and structural characterization ofTrichoderma reeseidehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Appl.Environ.Microbiol., 2019
|
|
3ZFP
 
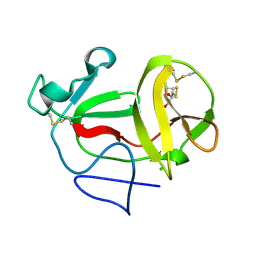 | | Crystal structure of product-like, processed N-terminal protease Npro with internal His-Tag | | 分子名称: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
6I3O
 
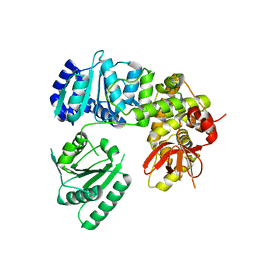 | |
3ZFT
 
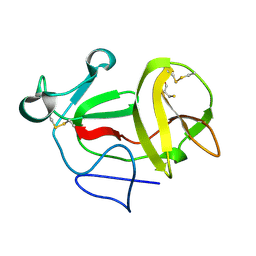 | | Crystal structure of product-like, processed N-terminal protease Npro at pH 3 | | 分子名称: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
7JNX
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)pivalamide | | 分子名称: | 2,2-dimethyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Andring, J.T, McKenna, R. | | 登録日 | 2020-08-05 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.286 Å) | | 主引用文献 | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
6I45
 
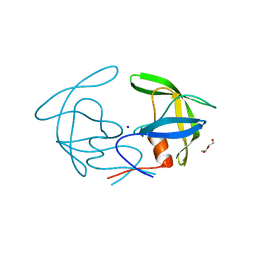 | | Crystal structure of I13V/I62V/V77I South African HIV-1 subtype C protease containing a D25A mutation | | 分子名称: | DI(HYDROXYETHYL)ETHER, Protease, SODIUM ION | | 著者 | Sherry, D, Pandian, R, Achilonu, I.A, Dirr, H.W, Sayed, Y. | | 登録日 | 2018-11-09 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Non-active site mutations in the HIV protease: Diminished drug binding affinity is achieved through modulating the hydrophobic sliding mechanism.
Int.J.Biol.Macromol., 217, 2022
|
|
7JO0
 
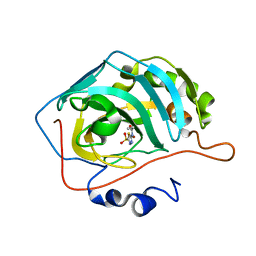 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propionamide | | 分子名称: | Carbonic anhydrase 2, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, ZINC ION | | 著者 | Andring, J.T, McKenna, R. | | 登録日 | 2020-08-05 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.607 Å) | | 主引用文献 | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
