4I3K
 
 | | Crystal structure of a metabolic reductase with 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one | | Descriptor: | 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, B, Yao, Y, Liu, Z, Deng, L, Anglin, J.L, Jiang, H, Prasad, B.V.V, Song, Y. | | Deposit date: | 2012-11-26 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3056 Å) | | Cite: | Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase.
ACS Med Chem Lett, 4, 2013
|
|
7LR9
 
 | | Crystal structure of KPC-2 S70G/T215P mutant with hydrolyzed imipenem | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of KPC-2 T215P mutant
To Be Published
|
|
8SJ3
 
 | | Beta-lactamase CTX-M-14 E166Y/N170G | | Descriptor: | Beta-lactamase CTX-M-14 | | Authors: | Judge, A, Palzkill, T, Sankaran, B, Prasad, B.V.V, Hu, L. | | Deposit date: | 2023-04-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Network of epistatic interactions in an enzyme active site revealed by large-scale deep mutational scanning.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SQF
 
 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
3QI0
 
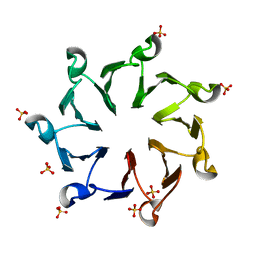 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase inhibitory protein II, SULFATE ION | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
3RAS
 
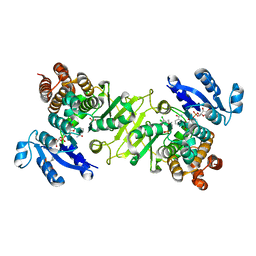 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with a lipophilic phosphonate inhibitor | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-(N-HYDROXYACETAMIDO)-1-(3,4-DICHLOROPHENYL)PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Diao, J, Deng, L, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies.
J.Med.Chem., 54, 2011
|
|
3QHY
 
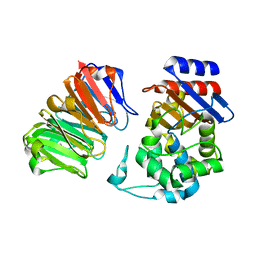 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase, Beta-lactamase inhibitory protein II | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
3SLN
 
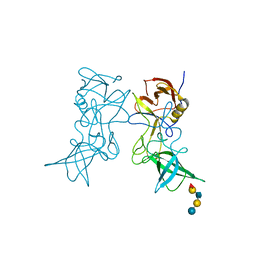 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to H pentasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SEJ
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to Secretor Lewis HBGA (LeB) | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SLD
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to A trisaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SKB
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SJP
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid, ZINC ION | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
6V6G
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6P
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | Deposit date: | 2019-11-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
5TWE
 
 | | CTX-M-14 P167S:S70G mutant enzyme crystallized with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
6V8V
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-2 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V83
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-1 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V5E
 
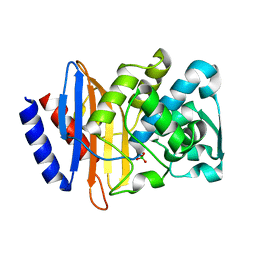 | | Crystal structure of CTX-M-14 P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V7T
 
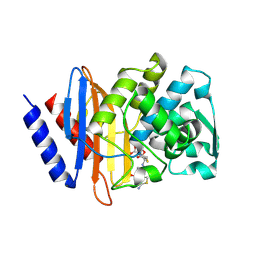 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase in complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6VP0
 
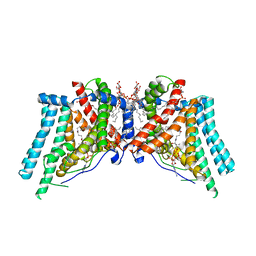 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
5TWD
 
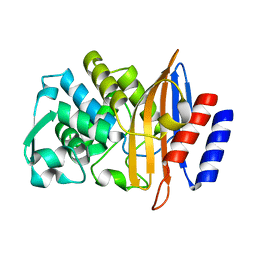 | | CTX-M-14 P167S apoenzyme | | Descriptor: | Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TW6
 
 | | CTX-M-14 P167S:E166A mutant with acylated ceftazidime molecule | | Descriptor: | 1,2-ETHANEDIOL, ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-11 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5U53
 
 | | CTX-M-14 E166A with acylated ceftazidime molecule | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase, NITRATE ION | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-12-06 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5VTH
 
 | | CTX-M-14 P167S:E166A mutant | | Descriptor: | Beta-lactamase | | Authors: | Hu, L, Patel, M, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2017-05-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
