6NIR
 
 | | Crystal structure of a GII.4 norovirus HOV protease | | Descriptor: | HOV protease, HOV protease fragment | | Authors: | Prasad, B.V.V, Hu, L. | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | GII.4 Norovirus Protease Shows pH-Sensitive Proteolysis with a Unique Arg-His Pairing in the Catalytic Site.
J. Virol., 93, 2019
|
|
4IMZ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN1
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, SULFATE ION | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMQ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN2
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
8TL1
 
 | |
8TKA
 
 | |
8TL8
 
 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | Descriptor: | GLYCOCHOLIC ACID, Protein sigma-NS | | Authors: | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
8G0W
 
 | |
7R6Z
 
 | | OXA-48 bound by Compound 3.3 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
3SR4
 
 | | Crystal Structure of Human DOT1L in Complex with a Selective Inhibitor | | Descriptor: | (2S)-2-azanyl-4-[[(2S,3S,4R,5R)-5-[6-(methylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Diao, J, Chen, P, Yao, Y, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-07-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective Inhibitors of Histone Methyltransferase DOT1L: Design, Synthesis, and Crystallographic Studies.
J.Am.Chem.Soc., 133, 2011
|
|
7UON
 
 | | CTX-M-14 Y105W mutant | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Judge, A, Hu, L, Sankaran, B, Van Riper, J, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2022-04-13 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mapping the determinants of catalysis and substrate specificity of the antibiotic resistance enzyme CTX-M beta-lactamase.
Commun Biol, 6, 2023
|
|
4WBA
 
 | | Q/E mutant SA11 NSP4_CCD | | Descriptor: | GLYCEROL, Non-structural glycoprotein NSP4, PHOSPHATE ION | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
7L8O
 
 | | OXA-48 bound by Compound 4.3 | | Descriptor: | 1,2-ETHANEDIOL, 9H-fluorene-2,7-disulfonate, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
2C86
 
 | | x-ray structure of the N and C-terminal domain of coronavirus nucleocapsid protein. | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collinson, E.W, Lescar, J, Prasad, B.V.V. | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation.
J.Virol., 80, 2006
|
|
2CA1
 
 | | Crystal structure of the IBV coronavirus nucleocapsid | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collison, E.W, Lescar, J, Prasad, B.V.V. | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation.
J.Virol., 80, 2006
|
|
3JYI
 
 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
4DS0
 
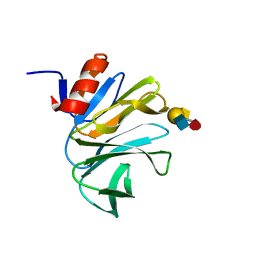 | | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hu, L, Crawford, S.E, Czako, R, Cortes-Penfield, N.W, Smith, D.F, Le Pendu, J, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen.
Nature, 485, 2012
|
|
4DRV
 
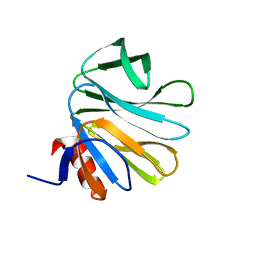 | | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Hu, L, Crawford, S.E, Czako, R, Cortes-Penfield, N.W, Smith, D.F, Le Pendu, J, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen.
Nature, 485, 2012
|
|
4DRR
 
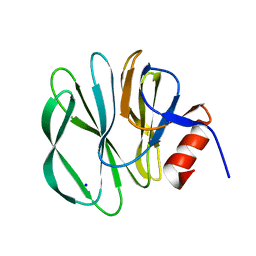 | | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen | | Descriptor: | Outer capsid protein VP4, SODIUM ION | | Authors: | Hu, L, Crawford, S.E, Czako, R, Cortes-Penfield, N.W, Smith, D.F, Le Pendu, J, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen.
Nature, 485, 2012
|
|
6XQR
 
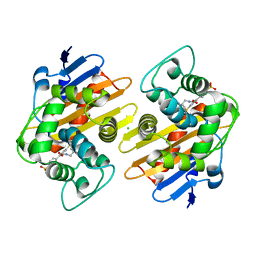 | | OXA-48 bound by Compound 2.2 | | Descriptor: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
5K4P
 
 | | Catalytic Domain of MCR-1 phosphoethanolamine transferase | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, sorbitol | | Authors: | Stojanoski, V, Palzkill, T, Prasad, B.V.V, Sankaran, B. | | Deposit date: | 2016-05-21 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Structure of the catalytic domain of the colistin resistance enzyme MCR-1.
Bmc Biol., 14, 2016
|
|
5KW9
 
 | |
