6NIR
 
 | | Crystal structure of a GII.4 norovirus HOV protease | | Descriptor: | HOV protease, HOV protease fragment | | Authors: | Prasad, B.V.V, Hu, L. | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | GII.4 Norovirus Protease Shows pH-Sensitive Proteolysis with a Unique Arg-His Pairing in the Catalytic Site.
J. Virol., 93, 2019
|
|
4IMZ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN1
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, SULFATE ION | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMQ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN2
 
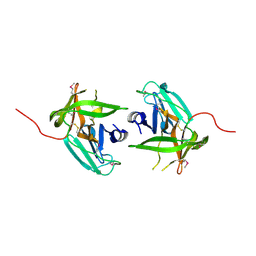 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
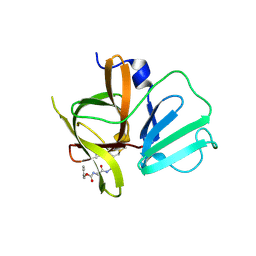 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
1IHM
 
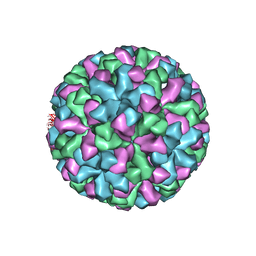 | | CRYSTAL STRUCTURE ANALYSIS OF NORWALK VIRUS CAPSID | | Descriptor: | capsid protein | | Authors: | Prasad, B.V, Hardy, M.E, Dokland, T, Bella, J, Rossmann, M.G, Estes, M.K. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray crystallographic structure of the Norwalk virus capsid
Science, 286, 1999
|
|
8TL1
 
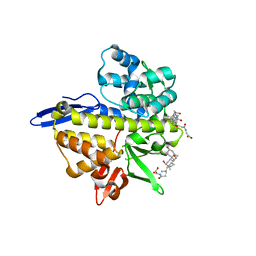 | |
8TKA
 
 | |
8TL8
 
 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | Descriptor: | GLYCOCHOLIC ACID, Protein sigma-NS | | Authors: | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
1UH8
 
 | |
1UH9
 
 | |
1UH7
 
 | |
7R6Z
 
 | | OXA-48 bound by Compound 3.3 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
8G0W
 
 | |
6AIE
 
 | | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution | | Descriptor: | Putative methyltransferase | | Authors: | Venkataraman, S, Dhankar, A, Sinha, K.M, Manivasakan, P, Iqbal, N, Singh, T.P, Prasad, B.V.L.S. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution
To Be Published
|
|
6AUK
 
 | |
7L8O
 
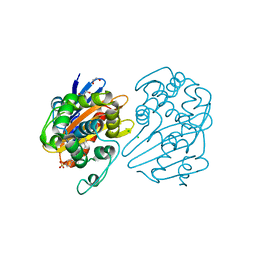 | | OXA-48 bound by Compound 4.3 | | Descriptor: | 1,2-ETHANEDIOL, 9H-fluorene-2,7-disulfonate, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
4WBA
 
 | | Q/E mutant SA11 NSP4_CCD | | Descriptor: | GLYCEROL, Non-structural glycoprotein NSP4, PHOSPHATE ION | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
3SR4
 
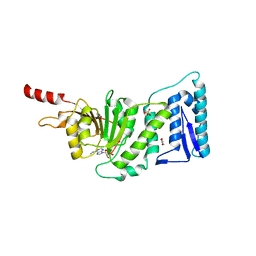 | | Crystal Structure of Human DOT1L in Complex with a Selective Inhibitor | | Descriptor: | (2S)-2-azanyl-4-[[(2S,3S,4R,5R)-5-[6-(methylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Diao, J, Chen, P, Yao, Y, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-07-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective Inhibitors of Histone Methyltransferase DOT1L: Design, Synthesis, and Crystallographic Studies.
J.Am.Chem.Soc., 133, 2011
|
|
6PCU
 
 | |
7RSW
 
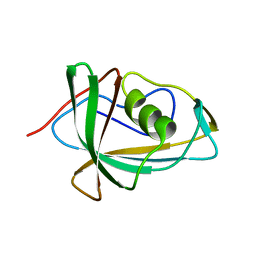 | | Crystal structure of group B human rotavirus VP8* | | Descriptor: | Outer capsid protein VP4, peptide | | Authors: | Hu, L, Salmen, W, Sankaran, B, Prasad, B.V. | | Deposit date: | 2021-08-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography.
Commun Biol, 5, 2022
|
|
2C86
 
 | | x-ray structure of the N and C-terminal domain of coronavirus nucleocapsid protein. | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collinson, E.W, Lescar, J, Prasad, B.V.V. | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation.
J.Virol., 80, 2006
|
|
2CA1
 
 | | Crystal structure of the IBV coronavirus nucleocapsid | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collison, E.W, Lescar, J, Prasad, B.V.V. | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation.
J.Virol., 80, 2006
|
|
