6NIR
 
 | |
1IHM
 
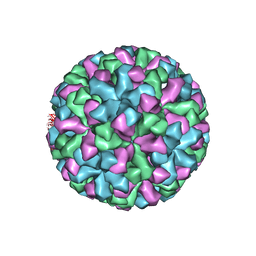 | | CRYSTAL STRUCTURE ANALYSIS OF NORWALK VIRUS CAPSID | | 分子名称: | capsid protein | | 著者 | Prasad, B.V, Hardy, M.E, Dokland, T, Bella, J, Rossmann, M.G, Estes, M.K. | | 登録日 | 2001-04-19 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | X-ray crystallographic structure of the Norwalk virus capsid
Science, 286, 1999
|
|
8TL8
 
 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | 分子名称: | GLYCOCHOLIC ACID, Protein sigma-NS | | 著者 | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | 登録日 | 2023-07-26 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
8TL1
 
 | |
8TKA
 
 | |
4IN1
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | 分子名称: | 3C-like protease, SULFATE ION | | 著者 | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | 登録日 | 2013-01-03 | | 公開日 | 2013-02-20 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMQ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | 分子名称: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | 著者 | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | 登録日 | 2013-01-03 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN2
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | 分子名称: | C-like protease | | 著者 | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | 登録日 | 2013-01-03 | | 公開日 | 2013-02-20 | | 最終更新日 | 2013-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | 分子名称: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | 著者 | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | 登録日 | 2013-01-04 | | 公開日 | 2013-02-20 | | 最終更新日 | 2013-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
1UH7
 
 | |
1UH9
 
 | |
1UH8
 
 | |
4IMZ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | 分子名称: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | 著者 | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | 登録日 | 2013-01-03 | | 公開日 | 2013-02-20 | | 最終更新日 | 2013-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4OPH
 
 | | X-ray structure of full-length H6N6 NS1 | | 分子名称: | Nonstructural protein 1 | | 著者 | Carrillo, B, Choi, J.M, Bornholdt, Z.A, Sankaran, S, Rice, A.P, Prasad, B.V.V. | | 登録日 | 2014-02-05 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.158 Å) | | 主引用文献 | The Influenza A Virus Protein NS1 Displays Structural Polymorphism.
J.Virol., 88, 2014
|
|
4YFZ
 
 | |
7RSW
 
 | | Crystal structure of group B human rotavirus VP8* | | 分子名称: | Outer capsid protein VP4, peptide | | 著者 | Hu, L, Salmen, W, Sankaran, B, Prasad, B.V. | | 登録日 | 2021-08-11 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography.
Commun Biol, 5, 2022
|
|
5VX5
 
 | | VP8* of a G2P[4] Human Rotavirus in complex with LNFP1 | | 分子名称: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Hu, L, Venkataram Prasad, B.V. | | 登録日 | 2017-05-23 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.285 Å) | | 主引用文献 | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
7R6Z
 
 | | OXA-48 bound by Compound 3.3 | | 分子名称: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | 登録日 | 2021-06-24 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
4F6Z
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | 分子名称: | Beta-lactamase, CITRATE ANION, ZINC ION | | 著者 | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2012-05-15 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
4FKD
 
 | | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta | | 分子名称: | Protein kinase C theta type, ZINC ION | | 著者 | Rahman, G.M, Shanker, S, Lewin, N.E, Prasad, B.V.V, Blumberg, P.M, Das, J. | | 登録日 | 2012-06-13 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.633 Å) | | 主引用文献 | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta.
Biochem.J., 451, 2013
|
|
5HAR
 
 | | OXA-163 beta-lactamase - S70G mutant | | 分子名称: | ACETATE ION, Beta-lactamase, CHLORIDE ION | | 著者 | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2015-12-30 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
5HAI
 
 | | P99 beta-lactamase mutant - S64G | | 分子名称: | Beta-lactamase, PHOSPHATE ION | | 著者 | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2015-12-30 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
7UON
 
 | | CTX-M-14 Y105W mutant | | 分子名称: | Beta-lactamase, PHOSPHATE ION | | 著者 | Judge, A, Hu, L, Sankaran, B, Van Riper, J, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2022-04-13 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Mapping the determinants of catalysis and substrate specificity of the antibiotic resistance enzyme CTX-M beta-lactamase.
Commun Biol, 6, 2023
|
|
7JHQ
 
 | | OXA-48 bound by Compound 2.3 | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase OXA-48, CHLORIDE ION, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-07-21 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6XQR
 
 | | OXA-48 bound by Compound 2.2 | | 分子名称: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-07-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
