5JEQ
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50K) in symmetric apo state | | Descriptor: | Nitrate/nitrite sensor protein NarQ, PHOSPHATE ION | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
4J39
 
 | |
5KDI
 
 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
8PMF
 
 | |
6WGD
 
 | | Crystal structure of a 6-phospho-beta-glucosidase from Bacillus licheniformis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2020-04-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structure, Bioinformatics Analysis, and Substrate Specificity of a 6-Phospho-beta-glucosidase Glycoside Hydrolase 1 Enzyme from Bacillus licheniformis .
J.Chem.Inf.Model., 60, 2020
|
|
4YZT
 
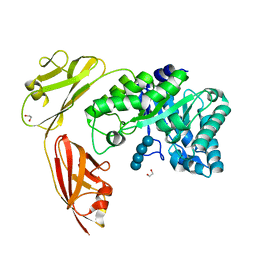 | | Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, Cellulose hydrolase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism.
Sci Rep, 6, 2016
|
|
4YZP
 
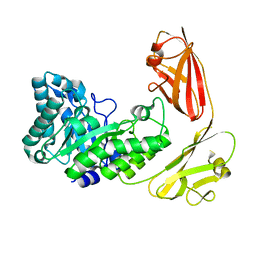 | |
5A44
 
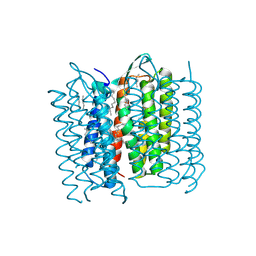 | | Structure of Bacteriorhodopsin obtained from 20um crystals by multi crystal data collection | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, HEPTANE, ... | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-05 | | Release date: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A3Y
 
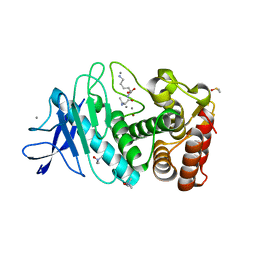 | | SAD structure of Thermolysin obtained by multi crystal data collection | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-04 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4KNQ
 
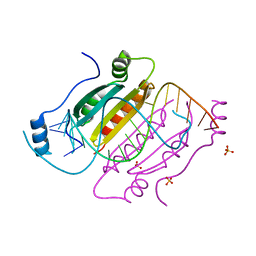 | |
5JGP
 
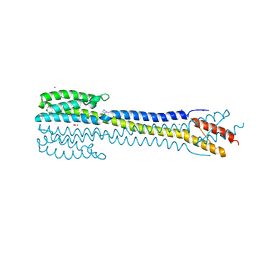 | | Crystal structure of the nitrate/nitrite sensor NarQ fragment bound with iodide ions | | Descriptor: | IODIDE ION, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Melnikov, I, Polovinkin, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
4KTG
 
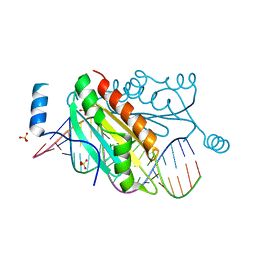 | |
5EDN
 
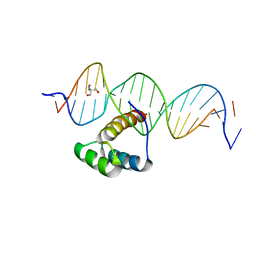 | | Structure of HOXB13-DNA(TCG) complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(P*GP*GP*AP*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
5EGO
 
 | | HOXB13-MEIS1 heterodimer bound to methylated DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5EEA
 
 | | Structure of HOXB13-DNA(CAA) complex | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
5EG0
 
 | | HOXB13-MEIS1 heterodimer bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Molecular basis of recognition of two distinct DNA sequences by a single transcription factor
To Be Published
|
|
8PMN
 
 | |
8PN4
 
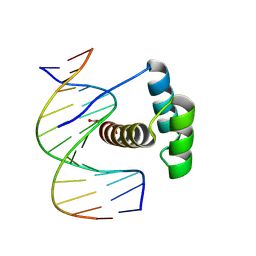 | |
8PNA
 
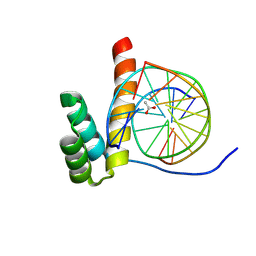 | |
5LHC
 
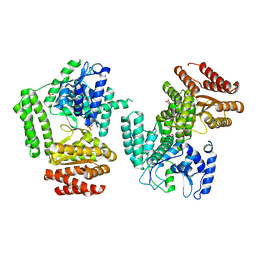 | | The structure of D456A mutant of Nt.BspD6I nicking endonuclease at 0.24 nm resolution . | | Descriptor: | GLYCEROL, Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Yunusova, A.K, Popov, A.N, Artyukh, R.I, Perevyazova, T.A, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural implication of activity loss by D456A mutant of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
6EYU
 
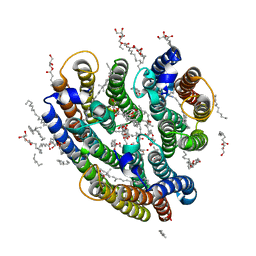 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
5JRF
 
 | | Crystal structure of the light-driven sodium pump KR2 bound with iodide ions | | Descriptor: | EICOSANE, IODIDE ION, Sodium pumping rhodopsin | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5JSI
 
 | | Structure of membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
6H24
 
 | |
5EF6
 
 | | Structure of HOXB13 complex with methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
