1QFE
 
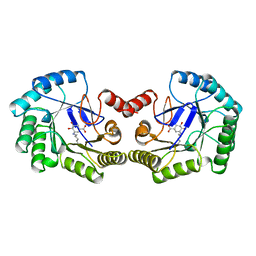 | | THE STRUCTURE OF TYPE I 3-DEHYDROQUINATE DEHYDRATASE FROM SALMONELLA TYPHI | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, PROTEIN (3-DEHYDROQUINATE DEHYDRATASE) | | Authors: | Shrive, A.K, Polikarpov, I, Sawyer, L, Coggins, J.R, Hawkins, A.R. | | Deposit date: | 1999-04-05 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction.
Nat.Struct.Biol., 6, 1999
|
|
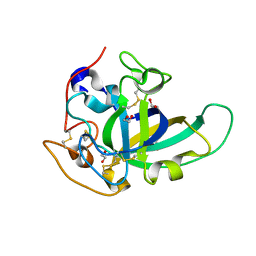
5KJO
 
 | |
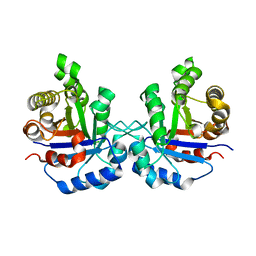
1R2S
 
 | |
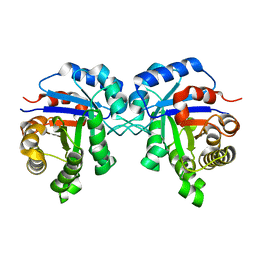
1R2T
 
 | |
1R8O
 
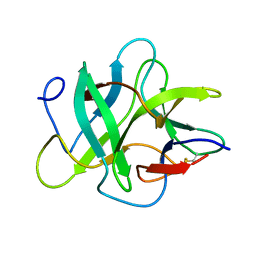 | | Crystal structure of an unusual Kunitz-type trypsin inhibitor from Copaifera langsdorffii seeds | | Descriptor: | Kunitz trypsin inhibitor | | Authors: | Krauchenco, S, Nagem, R.A.P, da Silva, J.A, Marangoni, S, Polikarpov, I. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Three-dimensional structure of an unusual Kunitz (STI) type trypsin inhibitor from Copaifera langsdorffii.
Biochimie, 86, 2004
|
|
1R8N
 
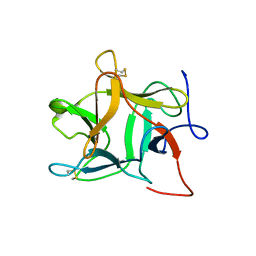 | |
3CFM
 
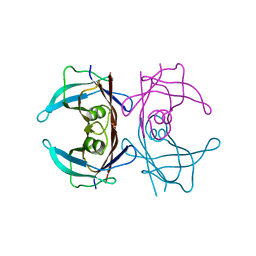 | |
3C9X
 
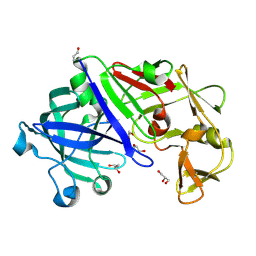 | | Crystal structure of Trichoderma reesei aspartic proteinase | | Descriptor: | GLYCEROL, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-02-19 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
3CFN
 
 | |
3CFT
 
 | |
3EMY
 
 | | Crystal structure of Trichoderma reesei aspartic proteinase complexed with pepstatin A | | Descriptor: | Pepstatin, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal
structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
1GMZ
 
 | |
3CFQ
 
 | |
3EXD
 
 | |
3KGU
 
 | |
3KGS
 
 | | V30M mutant human transthyretin (TTR) (apoV30M) pH 7.5 | | Descriptor: | Transthyretin | | Authors: | Trivella, D.B, Polikarpov, I. | | Deposit date: | 2009-10-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational differences between the wild type and V30M mutant transthyretin modulate its binding to genistein: implications to tetramer stability and ligand-binding.
J.Struct.Biol., 170, 2010
|
|
7K1R
 
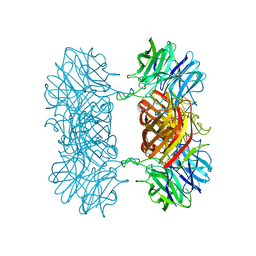 | |
7L1Z
 
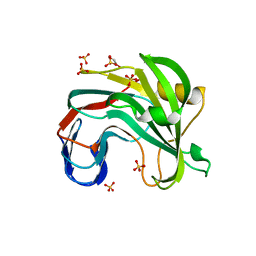 | | Unlocking the structural features for the exo-xylobiosidase activity of an unusual GH11 member identified in a compost-derived consortium - NT-truncated form | | Descriptor: | 1,2-ETHANEDIOL, Exo-B-1,4-beta-xylanase, SULFATE ION | | Authors: | Kadowaki, M.A.S, Polikarpov, I, Briganti, L, Evangelista, D.E. | | Deposit date: | 2020-12-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unlocking the structural features for the xylobiohydrolase activity of an unusual GH11 member identified in a compost-derived consortium.
Biotechnol.Bioeng., 118, 2021
|
|
7KW6
 
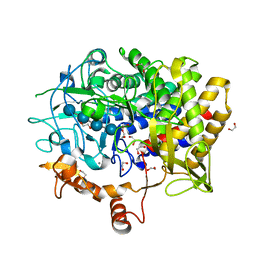 | | Crystal structure of the BlCel48B from Bacillus licheniformis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exoglucanase-2, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2020-11-30 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Impact of cellulose properties on enzymatic degradation by bacterial GH48 enzymes: Structural and mechanistic insights from processive Bacillus licheniformis Cel48B cellulase.
Carbohydr Polym, 264, 2021
|
|
3KGT
 
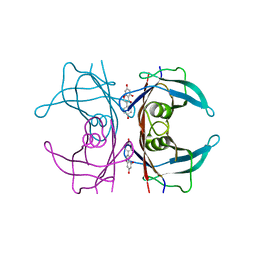 | |
3ILN
 
 | | X-ray structure of the laminarinase from Rhodothermus marinus | | Descriptor: | CALCIUM ION, GLYCEROL, Laminarinase | | Authors: | Bleicher, L, Golubev, A, Rojas, A.L, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of the thermostability and thermophilicity of laminarinases: X-ray structure of the hyperthermostable laminarinase from Rhodothermus marinus and molecular dynamics simulations.
J.Phys.Chem.B, 115, 2011
|
|
3HDL
 
 | | Crystal Structure of Highly Glycosylated Peroxidase from Royal Palm Tree | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Watanabe, L, Moura, P.R, Bleicher, L, Nascimento, A.S, Zamorano, L.S, Calvete, J.J, Bursakov, S, Roig, M.G, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2009-05-07 | | Release date: | 2009-11-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and statistical coupling analysis of highly glycosylated peroxidase from royal palm tree (Roystonea regia).
J.Struct.Biol., 169, 2010
|
|
3GWS
 
 | | Crystal Structure of T3-Bound Thyroid Hormone Receptor | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor beta | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Polikarpov, I, Baxter, J.D, Webb, P. | | Deposit date: | 2009-04-01 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function
J.Mol.Biol., 360, 2006
|
|
1QLL
 
 | | Piratoxin-II (Prtx-II) - a K49 PLA2 from Bothrops pirajai | | Descriptor: | N-TRIDECANOIC ACID, PHOSPHOLIPASE A2 | | Authors: | Lee, W.-H, Polikarpov, I. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Low Catalytic Activity in Lys49 Phospholipases A2-A Hypothesis: The Crystal Structure of Piratoxin II Complexed to Fatty Acid
Biochemistry, 40, 2001
|
|
3G9V
 
 | | Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22 | | Descriptor: | Interleukin 22 receptor, alpha 2, Interleukin-22 | | Authors: | de Moura, P.R, Watanabe, L, Bleicher, L, Colau, D, Renauld, J.-C, Polikarpov, I. | | Deposit date: | 2009-02-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22
FEBS Lett., 583, 2009
|
|
