5OC7
 
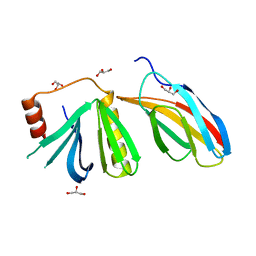 | | Crystal structure of the pleckstrin-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-PH_4). | | Descriptor: | Breakpoint cluster region protein,pleckstrin-homology domain of Bcr-Abl, D-MYO-INOSITOL-4,5-BISPHOSPHATE, GLYCEROL, ... | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-06-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
5N7E
 
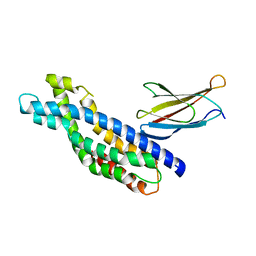 | | Crystal structure of the Dbl-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-DH_4). | | Descriptor: | Breakpoint cluster region protein, Mb(Bcr-DH_4) | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-02-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
8PI3
 
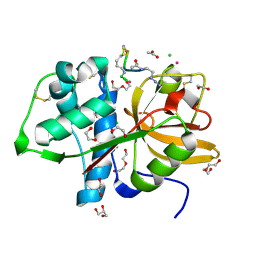 | | Cathepsin S Y132D mutant in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 2024
|
|
4MNX
 
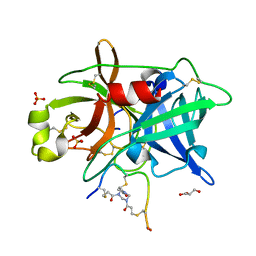 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK811 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8RND
 
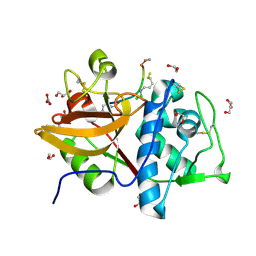 | | Cathepsin S in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin S, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 2024
|
|
4NCR
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with PBTZ169 | | Descriptor: | 2-(4-(cyclohexylmethyl)piperazin-1-yl)-8-nitro-6-(trifluoromethyl)-4H-benzo[e][1,3]thiazin-4-one, bound form, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2013-10-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Towards a new combination therapy for tuberculosis with next generation benzothiazinones.
EMBO Mol Med, 6, 2014
|
|
4MNW
 
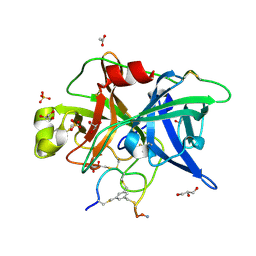 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK749 | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, ACETATE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MNY
 
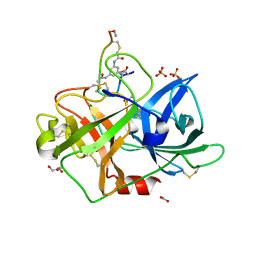 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK903 | | Descriptor: | ACETATE ION, GLYCEROL, N,N',N''-benzene-1,3,5-triyltris(2-bromoacetamide), ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3QF3
 
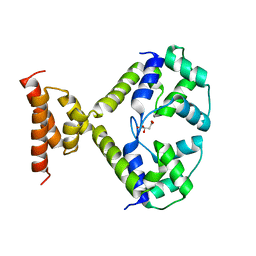 | |
3QYX
 
 | |
3R6S
 
 | |
3QWG
 
 | |
4OS5
 
 | |
4OS1
 
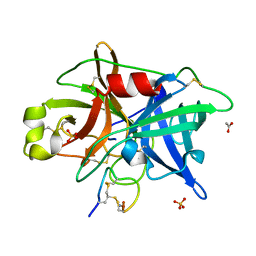 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK601 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS6
 
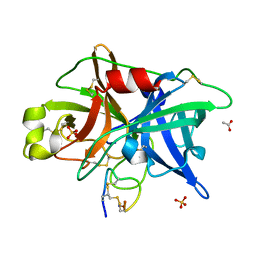 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK604 (bicyclic 2) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS4
 
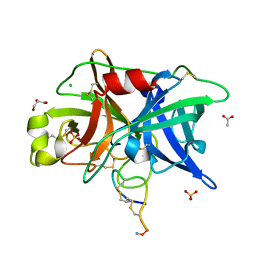 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK603 (bicyclic 1) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
7QO7
 
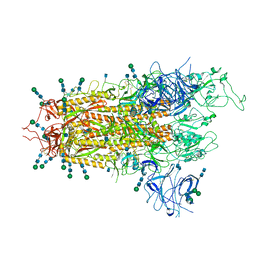 | | SARS-CoV-2 S Omicron Spike B.1.1.529 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
7QO9
 
 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD and NTD (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 S Omicron Spike B.1.1.529, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
7QUV
 
 | | Crystal structure of human Calprotectin (S100A8/S100A9) in complex with Peptide 3 | | Descriptor: | 1,2-ETHANEDIOL, 4-methanoyl-2-(6-oxidanyl-3-oxidanylidene-4~{H}-xanthen-9-yl)benzoic acid, AMINO GROUP, ... | | Authors: | Diaz-Perlas, C, Heinis, C, Pojer, F, Lau, K. | | Deposit date: | 2022-01-19 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-affinity peptides developed against calprotectin and their application as synthetic ligands in diagnostic assays.
Nat Commun, 14, 2023
|
|
7ZN1
 
 | | Avidin + Biotin-Tempo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(2,2,6,6-tetramethyl-1-oxidanyl-piperidin-4-yl)hexanamide, ... | | Authors: | Milani, J, Lau, K, Pojer, F, Ansermet, J.P, Saenz, F. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Avidin + Biotin-Tempo
To Be Published
|
|
8AQT
 
 | | Beta SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQU
 
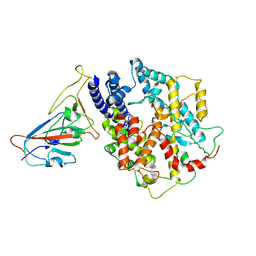 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQV
 
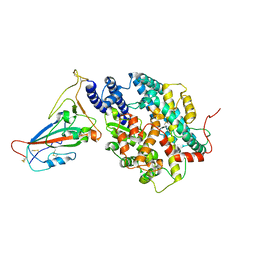 | | BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQS
 
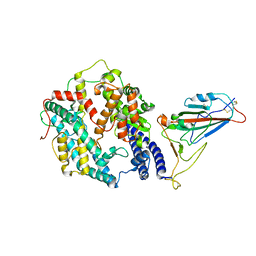 | | BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQW
 
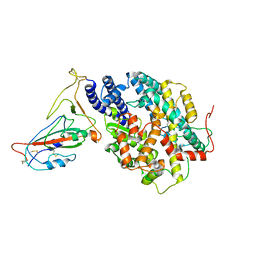 | | BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
