5I91
 
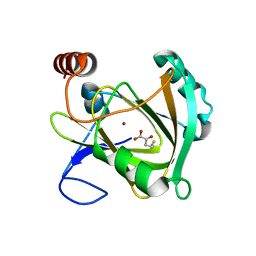 | | Structure of Mouse Acirecutone dioxygenase with to Ni2+ and 2-keto-4-(methylthio)-butyric acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, NICKEL (II) ION | | Authors: | Deshpande, A.R, Robinson, H, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I8S
 
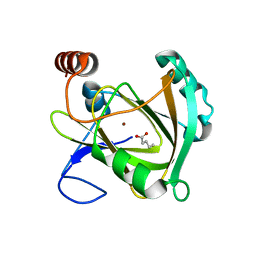 | | Structure of Mouse Acireductone dioxygenase with Ni2+ ion and pentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, NICKEL (II) ION, PENTANOIC ACID | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
4GCH
 
 | |
1YPI
 
 | |
2RGX
 
 | | Crystal Structure of Adenylate Kinase from Aquifex Aeolicus in complex with Ap5A | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Thai, V, Wolf-Watz, M, Fenn, T, Pozharski, E, Wilson, M.A, Petsko, G.A, Kern, D. | | Deposit date: | 2007-10-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intrinsic motions along an enzymatic reaction trajectory.
Nature, 450, 2007
|
|
3QYH
 
 | | Crystal Structure of Co-type Nitrile Hydratase beta-H71L from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QZ9
 
 | | Crystal structure of Co-type nitrile hydratase beta-Y215F from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
2RH5
 
 | | Structure of Apo Adenylate Kinase from Aquifex Aeolicus | | Descriptor: | Adenylate kinase | | Authors: | Thai, V, Wolf-Watz, M, Fenn, T, Pozharski, E, Wilson, M.A, Petsko, G.A, Kern, D. | | Deposit date: | 2007-10-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Intrinsic motions along an enzymatic reaction trajectory.
Nature, 450, 2007
|
|
3QYG
 
 | | Crystal Structure of Co-type Nitrile Hydratase beta-E56Q from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QZ5
 
 | | Crystal Structure of Co-type Nitrile Hydratase alpha-E168Q from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
5T1J
 
 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | Descriptor: | DNA, T-box transcription factor TBX21 | | Authors: | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3SDP
 
 | |
3CPO
 
 | | Crystal structure of ketosteroid isomerase D40N with bound 2-fluorophenol | | Descriptor: | 2-fluorophenol, Delta(5)-3-ketosteroid isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole
J.Am.Chem.Soc., 130, 2008
|
|
3BBG
 
 | |
1NXB
 
 | |
3FZW
 
 | | Crystal Structure of Ketosteroid Isomerase D40N-D103N from Pseudomonas putida (pKSI) with bound equilenin | | Descriptor: | EQUILENIN, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Ringe, D, Petsko, G.A. | | Deposit date: | 2009-01-26 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Hydrogen bond coupling in the ketosteroid isomerase active site.
Biochemistry, 48, 2009
|
|
3QXE
 
 | | Crystal Structure of Co-type Nitrile Hydratase from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
1DTN
 
 | |
1XYM
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | D-glucose, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
1XYA
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYL
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
1XYC
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | 3-O-METHYLFRUCTOSE IN LINEAR FORM, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYB
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | D-glucose, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
2R2D
 
 | | Structure of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, PHOSPHATE ION, ZINC ION, ... | | Authors: | Liu, D, Thomas, P.W, Momb, J, Hoang, Q, Petsko, G.A, Ringe, D, Fast, W. | | Deposit date: | 2007-08-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens.
Biochemistry, 46, 2007
|
|
3B35
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus | | Descriptor: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
