1UDI
 
 | |
1GOW
 
 | |
1QNL
 
 | |
1QO0
 
 | |
1UDH
 
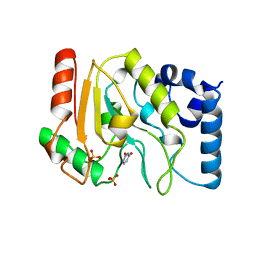 | |
1UDG
 
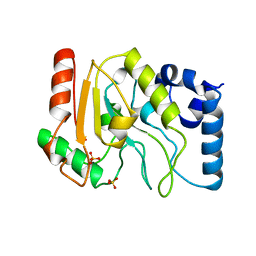 | |
1LAU
 
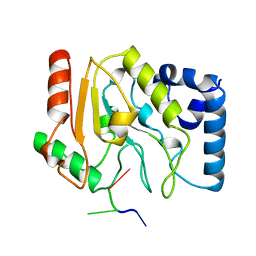 | | URACIL-DNA GLYCOSYLASE | | 分子名称: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE (E.C.3.2.2.-)) | | 著者 | Pearl, L.H, Savva, R. | | 登録日 | 1996-01-03 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Nature, 373, 1995
|
|
1PEA
 
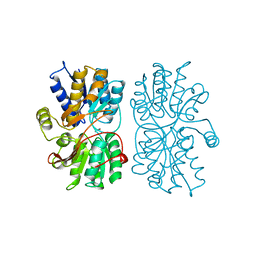 | |
1GXR
 
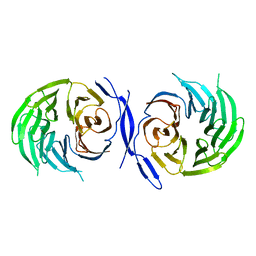 | | WD40 Region of Human Groucho/TLE1 | | 分子名称: | CALCIUM ION, TRANSDUCIN-LIKE ENHANCER PROTEIN 1 | | 著者 | Pearl, L.H, Roe, S.M, Pickles, L.M. | | 登録日 | 2002-04-10 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of the C-Terminal Wd40 Repeat Domain of the Human Groucho/Tle1 Transcriptional Corepressor
Structure, 10, 2002
|
|
4APE
 
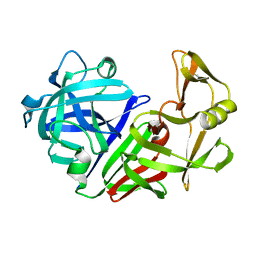 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | 分子名称: | ENDOTHIAPEPSIN | | 著者 | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | 登録日 | 1986-06-09 | | 公開日 | 1986-07-14 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
1AM1
 
 | |
1AMW
 
 | |
1W4S
 
 | |
6RML
 
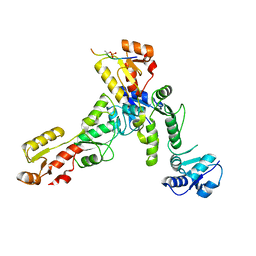 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a 53BP1 phosphopeptide | | 分子名称: | 53BP1, DNA topoisomerase 2-binding protein 1 | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2019-05-07 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint.
Elife, 8, 2019
|
|
8OK2
 
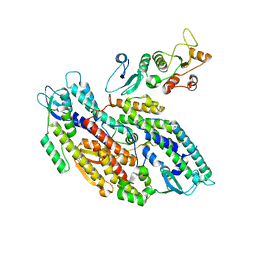 | | Bipartite interaction of TOPBP1 with the GINS complex | | 分子名称: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, DNA replication complex GINS protein PSF3, ... | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2023-03-26 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | TopBP1 utilises a bipartite GINS binding mode to support genome replication.
Nat Commun, 15, 2024
|
|
5HY6
 
 | |
6FO1
 
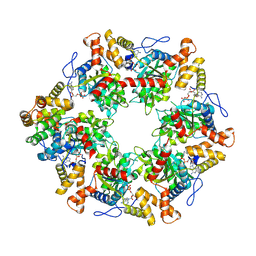 | | Human R2TP subcomplex containing 1 RUVBL1-RUVBL2 hexamer bound to 1 RBD domain from RPAP3. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RNA polymerase II-associated protein 3, RuvB-like 1, ... | | 著者 | Martino, F, Munoz-Hernandez, H, Rodriguez, C.F, Pearl, L.H, Llorca, O. | | 登録日 | 2018-02-05 | | 公開日 | 2018-04-04 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex.
Nat Commun, 9, 2018
|
|
5LOH
 
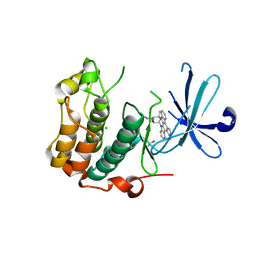 | | Kinase domain of human Greatwall | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, STAUROSPORINE, ... | | 著者 | Rajasekaran, M.B, Pearl, L.H, Oliver, A.W. | | 登録日 | 2016-08-09 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A first generation inhibitor of human Greatwall kinase, enabled by structural and functional characterisation of a minimal kinase domain construct.
Oncotarget, 7, 2016
|
|
7Z6H
 
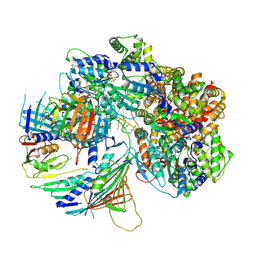 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | 分子名称: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2022-03-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
1MUG
 
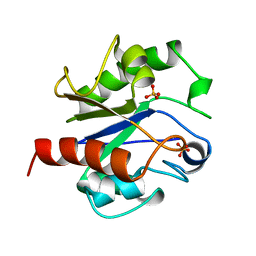 | | G:T/U MISMATCH-SPECIFIC DNA GLYCOSYLASE FROM E.COLI | | 分子名称: | PROTEIN (G:T/U SPECIFIC DNA GLYCOSYLASE), SULFATE ION | | 著者 | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | 登録日 | 1998-07-10 | | 公開日 | 1998-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1MWJ
 
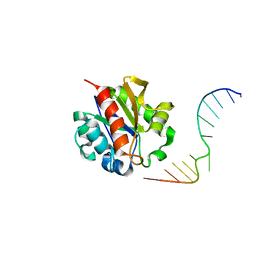 | | Crystal Structure of a MUG-DNA pseudo substrate complex | | 分子名称: | 5'-D(*CP*GP*CP*GP*A*GP*(DU)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | 著者 | Barrett, T.E, Scharer, O, Savva, R, Brown, T, Jiricny, J, Verdine, G.L, Pearl, L.H. | | 登録日 | 2002-09-30 | | 公開日 | 2002-10-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure of a thwarted mismatch glycosylase DNA repair complex
Embo J., 18, 1999
|
|
1MTL
 
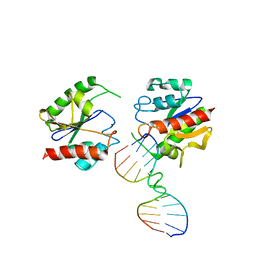 | | Non-productive MUG-DNA complex | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | 著者 | Barrett, T.E, Savva, R, Barlow, T, Brown, T, Jiricny, J, Pearl, L.H. | | 登録日 | 2002-09-21 | | 公開日 | 2002-09-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct.
Nat.Struct.Biol., 5, 1998
|
|
1MWI
 
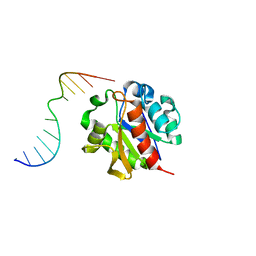 | | Crystal structure of a MUG-DNA product complex | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | 著者 | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | 登録日 | 2002-09-30 | | 公開日 | 2002-10-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
5ECG
 
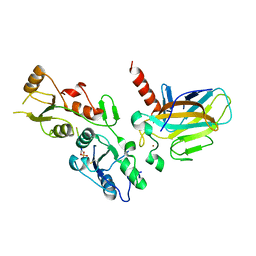 | | Crystal structure of the BRCT domains of 53BP1 in complex with p53 and H2AX-pSer139 (gammaH2AX) | | 分子名称: | Cellular tumor antigen p53, SEP-GLN-GLU-TYR, Tumor suppressor p53-binding protein 1, ... | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | ATM Localization and Heterochromatin Repair Depend on Direct Interaction of the 53BP1-BRCT2 Domain with gamma H2AX.
Cell Rep, 13, 2015
|
|
4UUH
 
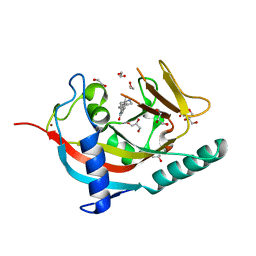 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 5-methyl-3-[4-(piperazin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, ... | | 著者 | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | 登録日 | 2014-07-28 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1- Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
