146D
 
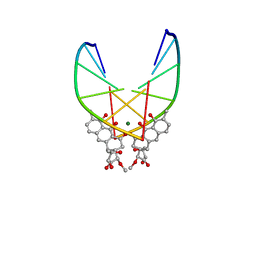 | | SOLUTION STRUCTURE OF THE MITHRAMYCIN DIMER-DNA COMPLEX | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, DNA (5'-D(*TP*CP*GP*CP*GP*A)-3'), ... | | Authors: | Sastry, M, Patel, D.J. | | Deposit date: | 1993-11-09 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mithramycin dimer-DNA complex.
Biochemistry, 32, 1993
|
|
1SLO
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLP
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, 16 STRUCTURES | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
5KDI
 
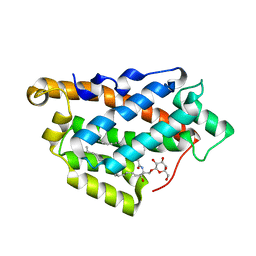 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
5KK5
 
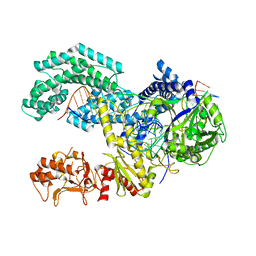 | | AsCpf1(E993A)-crRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cpf1, DNA (28-MER), DNA (8-mer), ... | | Authors: | Gao, P, Yang, H, Rajashankar, K.R, Huang, Z, Patel, D.J. | | Deposit date: | 2016-06-21 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.289 Å) | | Cite: | Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition.
Cell Res., 26, 2016
|
|
143D
 
 | |
3GL6
 
 | |
107D
 
 | | SOLUTION STRUCTURE OF THE COVALENT DUOCARMYCIN A-DNA DUPLEX COMPLEX | | Descriptor: | 4-HYDROXY-2,8-DIMETHYL-1-OXO-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-1,2,3,6,7,8-HEXAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*CP*CP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*GP*G)-3') | | Authors: | Lin, C.H, Patel, D.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the covalent duocarmycin A-DNA duplex complex.
J.Mol.Biol., 248, 1995
|
|
1A4T
 
 | | SOLUTION STRUCTURE OF PHAGE P22 N PEPTIDE-BOX B RNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | 20-MER BASIC PEPTIDE, BOXB RNA | | Authors: | Cai, Z, Gorin, A.A, Frederick, R, Ye, X, Hu, W, Majumdar, A, Kettani, A, Patel, D.J. | | Deposit date: | 1998-02-04 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex.
Nat.Struct.Biol., 5, 1998
|
|
1A6H
 
 | |
1AM0
 
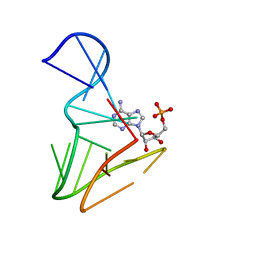 | | AMP RNA APTAMER COMPLEX, NMR, 8 STRUCTURES | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA APTAMER | | Authors: | Jiang, F, Kumar, R.A, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-06-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of RNA Folding and Recognition in an AMP-RNA Aptamer Complex
Nature, 382, 1996
|
|
1AFF
 
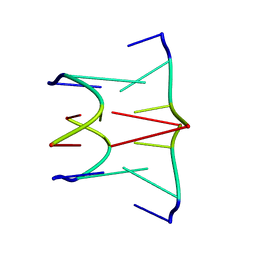 | | DNA QUADRUPLEX CONTAINING GGGG TETRADS AND (T.A).A TRIADS, NMR, 8 STRUCTURES | | Descriptor: | QUADRUPLEX DNA (5'-D(TP*AP*GP*G)-3') | | Authors: | Kettani, A, Bouaziz, S, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads.
Nat.Struct.Biol., 4, 1997
|
|
199D
 
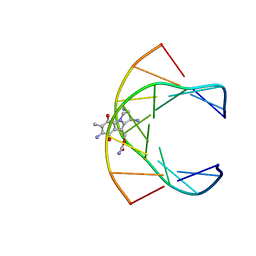 | | Solution structure of the monoalkylated mitomycin c-DNA complex | | Descriptor: | CARBAMIC ACID 2,6-DIAMINO-5-METHYL-4,7-DIOXO-2,3,4,7-TETRAHYDRO-1H-3A-AZA-CYCLOPENTA[A]INDEN-8-YLMETHYL ESTER, DNA (5'-D(*(DI)P*CP*AP*CP*GP*TP*CP*(DI)P*T)-3'), DNA (5'-D(*AP*CP*GP*AP*CP*GP*TP*GP*C)-3') | | Authors: | Sastry, M, Fiala, R, Lipman, R, Tomasz, M, Patel, D.J. | | Deposit date: | 1994-12-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monoalkylated mitomycin C-DNA complex.
J.Mol.Biol., 247, 1995
|
|
1A8N
 
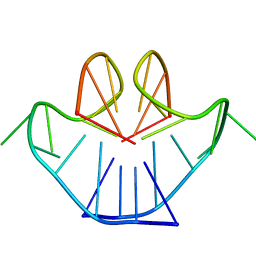 | | SOLUTION STRUCTURE OF A NA+ CATION STABILIZED DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS FORMED BY G-G-G-C REPEATS OBSERVED IN AAV AND HUMAN CHROMOSOME 19, NMR, 8 STRUCTURES | | Descriptor: | DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS | | Authors: | Kettani, A, Bouaziz, S, Gorin, A, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA.
J.Mol.Biol., 282, 1998
|
|
1B3P
 
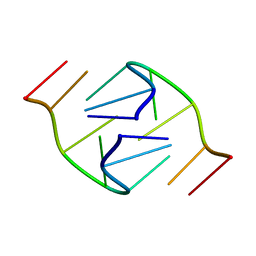 | | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*AP*T)-3') | | Authors: | Kettani, A, Bouaziz, S, Skripkin, E, Majumdar, A, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-12-14 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Interlocked mismatch-aligned arrowhead DNA motifs.
Structure Fold.Des., 7, 1999
|
|
1B5K
 
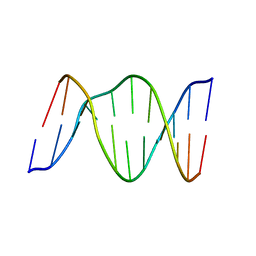 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1AW4
 
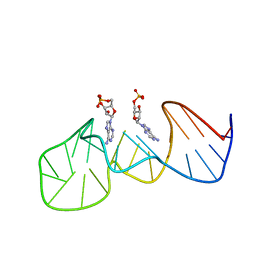 | |
1AX6
 
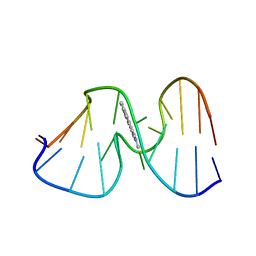 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT OPPOSITE A-2 DELETION SITE IN THE NARI HOT SPOT SEQUENCE CONTEXT; NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(CTCGGC-[AF]G-CCATC)D(GATGGCCGAG) | | Authors: | Mao, B, Gorin, A.A, Gu, Z, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a--2 deletion site in the NarI hot spot sequence context.
Biochemistry, 36, 1997
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1B6Y
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
1BIV
 
 | | BOVINE IMMUNODEFICIENCY VIRUS TAT-TAR COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | TAR RNA, TAT PEPTIDE | | Authors: | Ye, X, Kumar, R.A, Patel, D.J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex.
Chem.Biol., 2, 1995
|
|
1A8W
 
 | |
177D
 
 | |
1AXU
 
 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
