13GS
 
 | | GLUTATHIONE S-TRANSFERASE COMPLEXED WITH SULFASALAZINE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Ligandin (Non-Substrate) Binding Site of Human Pi Class Glutathione Transferase is Located in the Electrophile Binding Site (H-Site).
J.Mol.Biol., 291, 1999
|
|
4AHV
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(1H-pyrazol-1-yl)phenyl]methanamine, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
3TZR
 
 | | Structure of a Riboswitch-like RNA-ligand complex from the Hepatitis C Virus Internal Ribosome Entry Site | | Descriptor: | (8R)-8-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-1,7,8,9-tetrahydrochromeno[5,6-d]imidazol-2-amine, 5'-R(*CP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*CP*C)-3', 5'-R(*GP*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*UP*CP*GP*G)-3', ... | | Authors: | Dibrov, S.M, Ding, K, Brunn, N, Parker, M.A, Bergdahl, B.M, Wyles, D.L, Hermann, T. | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structure of a Riboswitch in the Hepatitis C Virus Internal Ribosome Entry Site
To be Published
|
|
3AO3
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 3-(1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO2
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-5-amine, ... | | Authors: | Wielens, J, Chalmers, D.K, Headey, S.J, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO5
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 5-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-3-amine, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO4
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 3-(1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-5-amine, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
1EKU
 
 | | CRYSTAL STRUCTURE OF A BIOLOGICALLY ACTIVE SINGLE CHAIN MUTANT OF HUMAN IFN-GAMMA | | Descriptor: | Interferon gamma, SULFATE ION | | Authors: | Landar, A, Curry, B, Parker, M.H, DiGiacomo, R, Indelicato, S.R, Walter, M.R. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, characterization, and structure of a biologically active single-chain mutant of human IFN-gamma.
J.Mol.Biol., 299, 2000
|
|
3BKC
 
 | | Crystal structure of anti-amyloid beta FAB WO2 (P21, FormB) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BKM
 
 | | Structure of anti-amyloid-beta Fab WO2 (Form A, P212121) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa, ... | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
1D5S
 
 | | CRYSTAL STRUCTURE OF CLEAVED ANTITRYPSIN POLYMER | | Descriptor: | P1-ARG ANTITRYPSIN | | Authors: | Dunstone, M.A, Dai, W, Whisstock, J.C, Rossjohn, J, Pike, R.N, Feil, S.C, Le Bonneic, B.F, Parker, M.W, Bottomley, S.P. | | Deposit date: | 1999-10-11 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cleaved antitrypsin polymers at atomic resolution.
Protein Sci., 9, 2000
|
|
3BKJ
 
 | | Crystal structure of Fab wo2 bound to the n terminal domain of amyloid beta peptide (1-16) | | Descriptor: | Amyloid Beta Peptide, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
1EGJ
 
 | | DOMAIN 4 OF THE BETA COMMON CHAIN IN COMPLEX WITH AN ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (HEAVY CHAIN), ANTIBODY (LIGHT CHAIN), ... | | Authors: | Rossjohn, J, McKinstry, W.J, Woodcock, J.M, McClure, B.J, Hercus, T.R, Parker, M.W, Lopez, A.F, Bagley, C.J. | | Deposit date: | 2000-02-15 | | Release date: | 2001-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the activation domain of the GM-CSF/IL-3/IL-5 receptor common beta-chain bound to an antagonist.
Blood, 95, 2000
|
|
2GP8
 
 | | NMR SOLUTION STRUCTURE OF THE COAT PROTEIN-BINDING DOMAIN OF BACTERIOPHAGE P22 SCAFFOLDING PROTEIN | | Descriptor: | PROTEIN (SCAFFOLDING PROTEIN) | | Authors: | Sun, Y, Parker, M.H, Weigele, P, Casjens, S, Prevelige Jr, P.E, Krishna, N.R. | | Deposit date: | 1999-05-11 | | Release date: | 1999-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus.
J.Mol.Biol., 297, 2000
|
|
3AKN
 
 | |
3AO1
 
 | | Fragment-based approach to the design of ligands targeting a novel site in HIV-1 integrase | | Descriptor: | 1,3-benzodioxol-5-ol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
1OQX
 
 | | G-2 glycovariant of human IgG Fc bound to minimized version of Protein A called Z34C | | Descriptor: | Protein A Z34C, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Raju, T.S, Mulkerrin, M.G, Parker, M, De Vos, A.M, Gazzano-Santoro, H, Totpal, K, Ultsch, M.H. | | Deposit date: | 2003-03-11 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Impact of Fc Glycans on The Effector Functions Vary
with the Antibody mechanism of Action
To be Published
|
|
4P8Q
 
 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Alanine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucyl-cystinyl aminopeptidase, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
11GS
 
 | | Glutathione s-transferase complexed with ethacrynic acid-glutathione conjugate (form ii) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Mazzetti, A.P, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The glutathione conjugate of ethacrynic acid can bind to human pi class glutathione transferase P1-1 in two different modes.
FEBS Lett., 419, 1997
|
|
16GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
4PJ6
 
 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Lysine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSINE, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
1OQO
 
 | | Complex between G0 version of an Fc bound to a minimized version of Protein A called Mini-Z | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Minimized version of Protein A (Z34C), ... | | Authors: | Raju, T.S, Mulkerrin, M.G, Parker, M, De Vos, A.M, Gazzano-Santoro, H, Totpal, K, Ultsch, M.H. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impact of Fc Glycans on The Effector
Functions Vary with the Antibody
mechanism of Action
To be Published
|
|
14GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
17GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glutathione S-transferase P1-1
To be published
|
|
3BAE
 
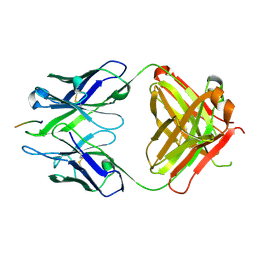 | | Crystal structure of Fab WO2 bound to the N terminal domain of Amyloid beta peptide (1-28) | | Descriptor: | Amyloid Beta Peptide, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Parker, M.W. | | Deposit date: | 2007-11-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
