7P9V
 
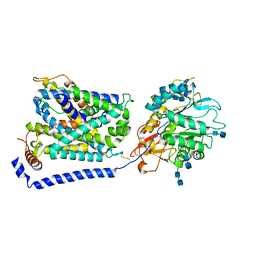 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9U
 
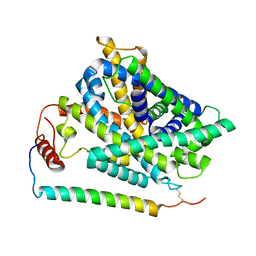 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7BC6
 
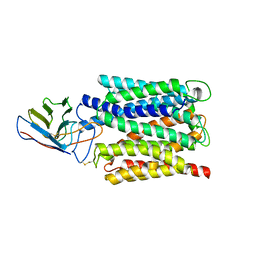 | | Cryo-EM structure of the outward open proton coupled folate transporter at pH 7.5 | | Descriptor: | Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
7BC7
 
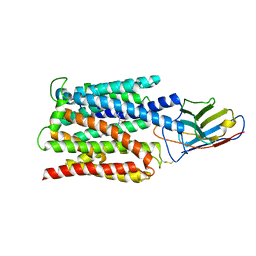 | | Cryo-EM structure of the proton coupled folate transporter at pH 6.0 bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
7NQK
 
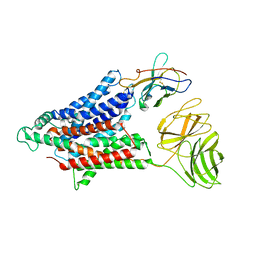 | | Cryo-EM structure of the mammalian peptide transporter PepT2 | | Descriptor: | Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-03-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of PepT2 reveals structural basis for proton-coupled peptide and prodrug transport in mammals.
Sci Adv, 7, 2021
|
|
9BIT
 
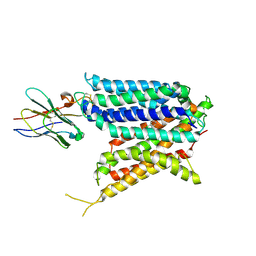 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cloxacillin, pose 1 | | Descriptor: | CLOXACILLIN, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIU
 
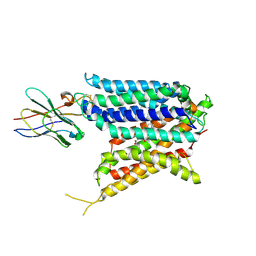 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cloxacillin, pose 2 | | Descriptor: | CLOXACILLIN, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIS
 
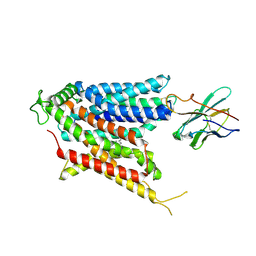 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
9BIR
 
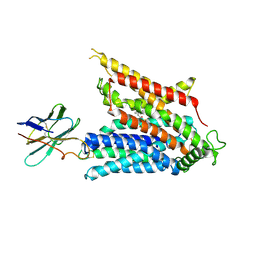 | | Cryo-EM structure of the mammalian peptide transporter PepT2 bound to cefadroxil | | Descriptor: | Cefadroxil, Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2024-04-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic transport and inhibition in PepT2.
Nat Commun, 15, 2024
|
|
7ZKW
 
 | |
8APY
 
 | | Crystal structure of the H12A variant of the KDEL receptor bound to sybody | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ER lumen protein-retaining receptor 2, Synthetic nanobody | | Authors: | Parker, J.L, Smith, K, Newstead, S. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-23 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Molecular basis for pH sensing in the KDEL trafficking receptor.
Structure, 32, 2024
|
|
8BVS
 
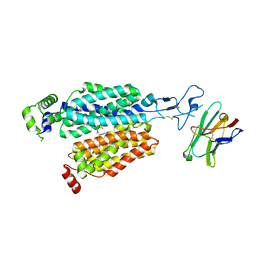 | | Cryo-EM structure of rat SLC22A6 bound to tenofovir | | Descriptor: | CHLORIDE ION, Solute carrier family 22 member 6, Synthetic nanobody (Sybody), ... | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BVR
 
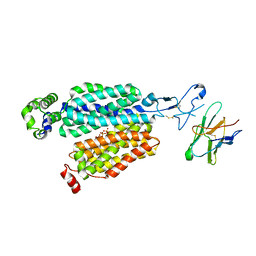 | | Cryo-EM structure of rat SLC22A6 in the apo state | | Descriptor: | PHOSPHATE ION, Solute carrier family 22 member 6, Synthetic nanobody (Sybody) | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BVT
 
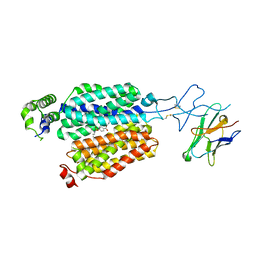 | | Cryo-EM structure of rat SLC22A6 bound to probenecid | | Descriptor: | 4-(dipropylsulfamoyl)benzoic acid, Solute carrier family 22 member 6, Synthetic nanobody (Sybody) | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BW7
 
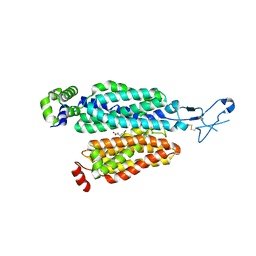 | | Cryo-EM structure of rat SLC22A6 bound to alpha-ketoglutaric acid | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Solute carrier family 22 member 6, ... | | Authors: | Parker, J.L, Kato, T, Newstead, S. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Molecular basis for selective uptake and elimination of organic anions in the kidney by OAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8P6A
 
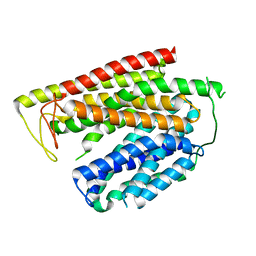 | |
8OMU
 
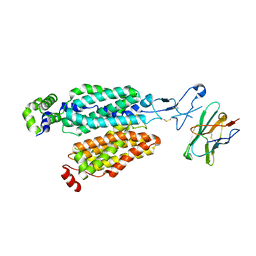 | |
5A2N
 
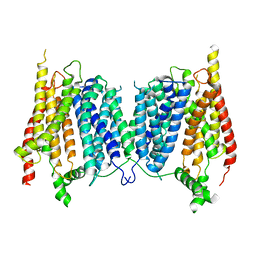 | |
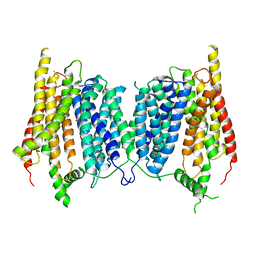
5A2O
 
 | |
7OXE
 
 | |
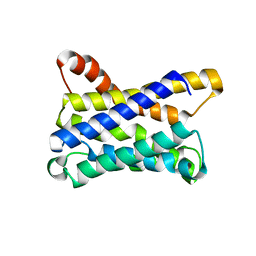
6ZXR
 
 | |
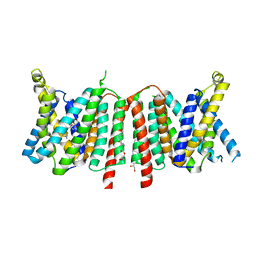
6QSK
 
 | |
7NVH
 
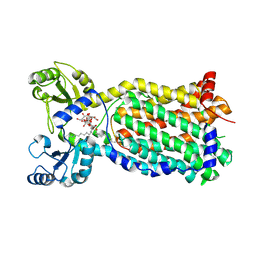 | | Cryo-EM structure of the mycolic acid transporter MmpL3 from M. tuberculosis | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Trehalose monomycolate exporter MmpL3 | | Authors: | Adams, O, Deme, J.C, Parker, J.L, Lea, S.M, Newstead, S. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure and resistance landscape of M. tuberculosis MmpL3: An emergent therapeutic target.
Structure, 29, 2021
|
|
5OGK
 
 | |
5OGE
 
 | |
