6W1J
 
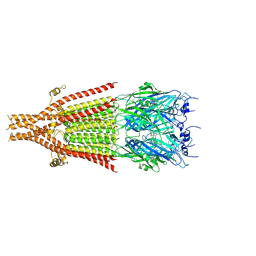 | | Cryo-EM structure of 5HT3A receptor in presence of Alosetron | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2020-03-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structures of multiple 5-HT 3A R-setron complexes reveal a novel mechanism of competitive inhibition.
Elife, 9, 2020
|
|
1PZM
 
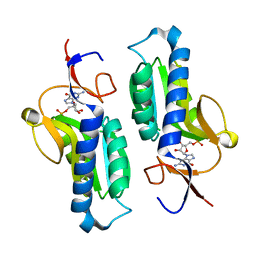 | | Crystal structure of HGPRT-ase from Leishmania tarentolae in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Monzani, P.S, Trapani, S, Oliva, G, Thiemann, O.H. | | Deposit date: | 2003-07-11 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Leishmania tarentolae hypoxanthine-guanine phosphoribosyltransferase.
Bmc Struct.Biol., 7, 2007
|
|
7N3V
 
 | | Crystal structure of Mycobacterium smegmatis LmcA | | Descriptor: | GLYCEROL, LmcA, SULFATE ION | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the putative cell-wall lipoglycan biosynthesis protein LmcA from Mycobacterium smegmatis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4X6S
 
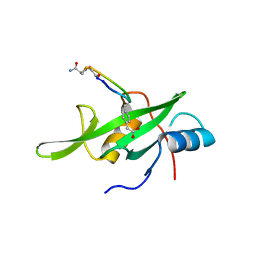 | | Grb7 SH2 domain with phosphotyrosine mimetic inhibitor peptide | | Descriptor: | Growth factor receptor-bound protein 7, Phosphotyrosine mimetic inhibitor peptide G7-TEM1 | | Authors: | Watson, G.M, Panjikar, S, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-09-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
4FXA
 
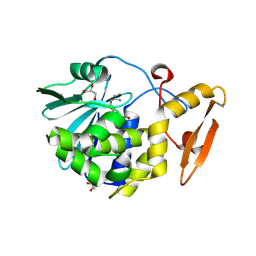 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl arginine at 1.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, N-ALPHA-L-ACETYL-ARGININE, ... | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-07-03 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl arginine at 1.7 Angstrom resolution
To be Published
|
|
1NWA
 
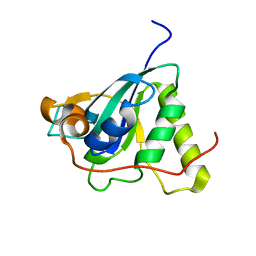 | | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Taylor, A.B, Benglis Jr, D.M, Dhandayuthapani, S, Hart, P.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine
J.Bacteriol., 185, 2003
|
|
4WVM
 
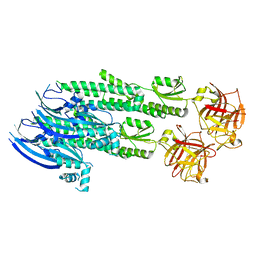 | | Stonustoxin structure | | Descriptor: | Stonustoxin subunit alpha, Stonustoxin subunit beta | | Authors: | Ellisdon, A.M, Panjikar, S, Whisstock, J.C, McGowan, S. | | Deposit date: | 2014-11-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stonefish toxin defines an ancient branch of the perforin-like superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6UE0
 
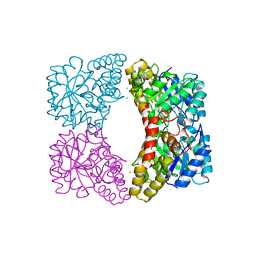 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
5VFY
 
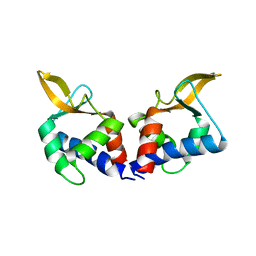 | | Structure of an accessory protein of the pCW3 relaxosome | | Descriptor: | TcpK | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
1YS7
 
 | | Crystal structure of the response regulator protein prrA complexed with Mg2+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The structural basis of signal transduction for the response regulator PrrA from Mycobacterium tuberculosis.
J.Biol.Chem., 281, 2006
|
|
1YS6
 
 | |
4J1Y
 
 | | The X-ray crystal structure of human complement protease C1s zymogen | | Descriptor: | Complement C1s subcomponent | | Authors: | Perry, A.J, Wijeyewickrema, L.C, Wilmann, P.G, Gunzburg, M.J, D'Andrea, L, Irving, J.A, Pang, S.S, Duncan, R.C, Wilce, J.A, Whisstock, J.C, Pike, R.N. | | Deposit date: | 2013-02-03 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6645 Å) | | Cite: | A Molecular Switch Governs the Interaction between the Human Complement Protease C1s and Its Substrate, Complement C4.
J.Biol.Chem., 288, 2013
|
|
4OQO
 
 | | Crystal structure of the tryptic generated iron-free C-lobe of bovine Lactoferrin at 2.42 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lactotransferrin | | Authors: | Singh, A, Rastogi, N, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the iron-free true C-terminal half of bovine lactoferrin produced by tryptic digestion and its functional significance in the gut.
Febs J., 281, 2014
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
4XG1
 
 | | Psychromonas ingrahamii diaminopimelate decarboxylase with LLP | | Descriptor: | (2S)-2-amino-6-[[3-hydroxy-2-methyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]hexanoic acid, Diaminopimelate decarboxylase, POTASSIUM ION, ... | | Authors: | Peverelli, M.G, Wubben, J.M, Panjikar, S, Perugini, M.A. | | Deposit date: | 2014-12-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Expression to crystallization of diaminopimelate decarboxylase from the psychrophile Psychromonas ingrahamii
To Be Published
|
|
1L0Z
 
 | | THE STRUCTURE OF PORCINE PANCREATIC ELASTASE COMPLEXED WITH XENON AND BROMIDE, CRYOPROTECTED WITH DRY PARAFFIN OIL | | Descriptor: | BROMIDE ION, ELASTASE 1, SODIUM ION, ... | | Authors: | Tucker, P.A, Panjikar, S. | | Deposit date: | 2002-02-14 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenon derivatization of halide-soaked protein crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4GUW
 
 | | Crystal structure of type 1 Ribosome inactivating protein from Momordica balsamina with lipopolysaccharide at 1.6 Angstrom resolution | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of type 1 Ribosome inactivating protein from Momordica balsamina with lipopolysaccharide at 1.6 Angstrom resolution
To be published
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
5IRA
 
 | | Expanding Nature's Catalytic Repertoire -Directed Evolution of an Artificial Metalloenzyme for In Vivo Metathesis | | Descriptor: | Artificial Metathesase, [1-[4-[[5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]methyl]-2,6-dimethyl-phenyl]-3-(2,4,6-trimethylphenyl)-4,5-dihydroimidazol-1-ium-2-yl]-bis(chloranyl)ruthenium | | Authors: | Heinisch, T, Jeschek, M, Reuter, R, Trindler, C, Panke, S, Ward, T.R. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Directed evolution of artificial metalloenzymes for in vivo metathesis.
Nature, 537, 2016
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
1LKB
 
 | | Porcine Pancreatic Elastase/Na-Complex | | Descriptor: | CHLORIDE ION, Elastase 1, SODIUM ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Nowak, E, Tucker, P.A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding to porcine pancreatic elastase: calcium or not calcium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
5VE6
 
 | | Crystal structure of Sugen kinase 223 | | Descriptor: | Tyrosine-protein kinase SgK223 | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association.
Nat Commun, 8, 2017
|
|
2WFL
 
 | | Crystal structure of polyneuridine aldehyde esterase | | Descriptor: | POLYNEURIDINE-ALDEHYDE ESTERASE, SULFATE ION | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5XAZ
 
 | |
