5NO1
 
 | |
5MMA
 
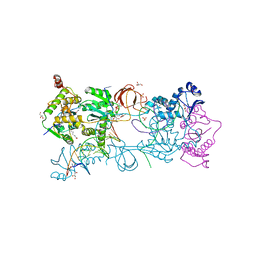 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ379 (compound 5'g) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors.
J. Med. Chem., 60, 2017
|
|
5ME9
 
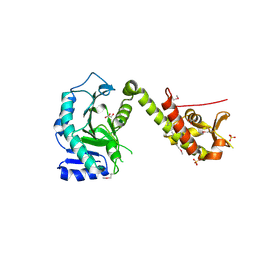 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 1. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5M0R
 
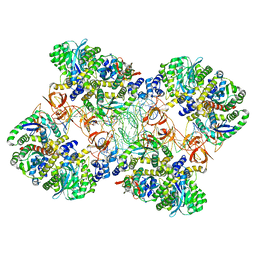 | | Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex | | Descriptor: | integrase, tDNA, vDNA, ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Maskell, D, Locke, J, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration.
Science, 355, 2017
|
|
5LLJ
 
 | |
5MEC
 
 | |
5MEA
 
 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 2. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5MEB
 
 | | Crystal structure of yeast Cdt1 C-terminal domain | | Descriptor: | Cell division cycle protein CDT1, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5M9U
 
 | |
6FMB
 
 | |
5CZ2
 
 | | Crystal structure of a two-domain fragment of MMTV integrase | | Descriptor: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | Authors: | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
1XX9
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with EcotinM84R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1ZPB
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 4-Methyl-pentanoic acid {1-[4-guanidino-1-(thiazole-2-carbonyl)-butylcarbamoyl]-2-methyl-propyl}-amide | | Descriptor: | 4-METHYL-PENTANOIC ACID {1-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYLCARBAMOYL]-2-METHYL-PROPYL}-AMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1XXD
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with mutated Ecotin | | Descriptor: | Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1XXF
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with Ecotin Mutant (EcotinP) | | Descriptor: | Coagulation factor XI, Ecotin, SODIUM ION | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1ZJD
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Kunitz Protease Inhibitor Domain of Protease Nexin II | | Descriptor: | Catalytic Domain of Coagulation Factor XI, Kunitz Protease Inhibitory Domain of Protease Nexin II | | Authors: | Jin, L, Navaneetham, D, Pandey, P, Strickler, J.E, Babine, R.E, Walsh, P.N, Abdel-Meguid, S.S. | | Deposit date: | 2005-04-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mutational Analyses of the Molecular Interactions between the Catalytic Domain of Factor XIa and the Kunitz Protease Inhibitor Domain of Protease Nexin 2
J.Biol.Chem., 280, 2005
|
|
1ZHR
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Benzamidine (S434A-T475A-C482S-K437A Mutant) | | Descriptor: | BENZAMIDINE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZHP
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Benzamidine (S434A-T475A-K505 Mutant) | | Descriptor: | BENZAMIDINE, GLUTATHIONE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZHM
 
 | | Crystal Structure of the Catalytic Domain of the Coagulation Factor XIa in Complex with Benzamidine (S434A-T475A-K437 Mutant) | | Descriptor: | BENZAMIDINE, GLUTATHIONE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZPC
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-[2-(3-Chloro-phenyl)-2-hydroxy-acetylamino]-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-3-methyl-butyramide | | Descriptor: | 2-[2-(3-CHLORO-PHENYL)-2-HYDROXY-ACETYLAMINO]-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-3-METHYL-BUTYRAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ZOM
 
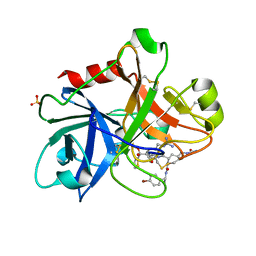 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in complex with a peptidomimetic Inhibitor | | Descriptor: | (S)-2-(3-((R)-1-(4-BROMOPHENYL)ETHYL)UREIDO)-N-((S)-1-((S)-5-GUANIDINO-1-OXO-1-(THIAZOL-2-YL)PENTAN-2-YLAMINO)-3-METHYL-1-OXOBUTAN-2-YL)-5-UREIDOPENTANAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Lin, J, Deng, H, Jin, L, Pandey, P, Rynkiewicz, M, Bibbins, F, Cantin, S, Quinn, J, Magee, S, Gorga, J. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, synthesis, and biological evaluation of peptidomimetic inhibitors of factor XIa as novel anticoagulants.
J.Med.Chem., 49, 2006
|
|
3WJ8
 
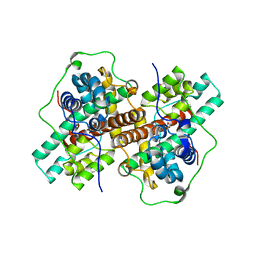 | | Crystal Structure of DL-2-haloacid dehalogenase mutant with 2-bromo-2-methylpropionate | | Descriptor: | 2-bromo-2-methylpropanoic acid, DL-2-haloacid dehalogenase, GLYCEROL | | Authors: | Siwek, A, Omi, R, Hirotsu, K, Jitsumori, K, Esaki, N, Kurihara, T, Paneth, P. | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding modes of DL-2-haloacid dehalogenase revealed by crystallography, modeling and isotope effects studies.
Arch.Biochem.Biophys., 540, 2013
|
|
2A1D
 
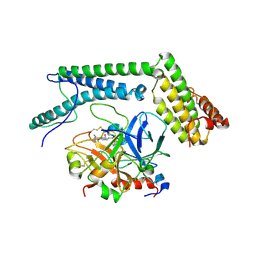 | | Staphylocoagulase bound to bovine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Friedrich, R, Panizzi, P, Kawabata, S, Bode, W, Bock, P.E, Fuentes-Prior, P. | | Deposit date: | 2005-06-20 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis for Reduced Staphylocoagulase-mediated Bovine Prothrombin Activation
J.Biol.Chem., 281, 2006
|
|
5CO4
 
 | |
1U59
 
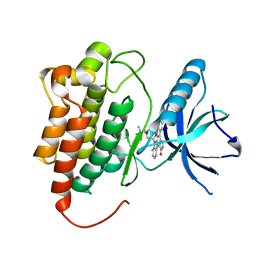 | | Crystal Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ZAP-70 | | Authors: | Jin, L, Pluskey, S, Petrella, E.C, Cantin, S.M, Gorga, J.C, Rynkiewicz, M.J, Pandey, P, Strickler, J.E, Babine, R.E, Weaver, D.T, Seidl, K.J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine: IMPLICATIONS FOR THE DESIGN OF SELECTIVE INHIBITORS
J.Biol.Chem., 279, 2004
|
|
