5CPF
 
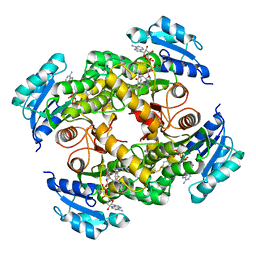 | | Compensation of the effect of isoleucine to alanine mutation by designed inhibition in the InhA enzyme | | 分子名称: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Li, H.-J, Lai, C.-T, Pan, P, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | 登録日 | 2015-07-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.409 Å) | | 主引用文献 | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
7U32
 
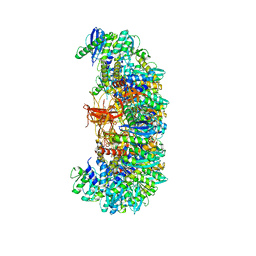 | | MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution | | 分子名称: | CALCIUM ION, DNA EV272, DNA EV273, ... | | 著者 | Shan, Z, Pye, V.E, Cherepanov, P, Lyumkis, D. | | 登録日 | 2022-02-25 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
7OUG
 
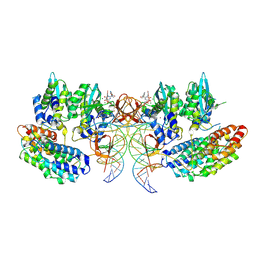 | | STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Integrase, ... | | 著者 | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | 登録日 | 2021-06-11 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUH
 
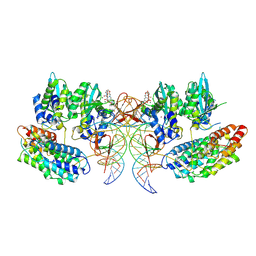 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir | | 分子名称: | Bictegravir, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | 著者 | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | 登録日 | 2021-06-11 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUF
 
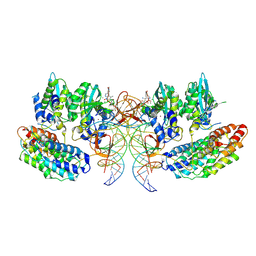 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450 | | 分子名称: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[3-(dimethylamino)-3-oxidanylidene-propyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | 著者 | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | 登録日 | 2021-06-11 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7NT9
 
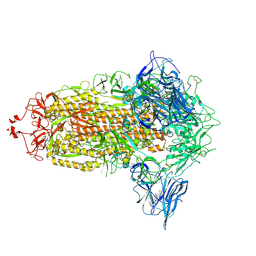 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | 登録日 | 2021-03-09 | | 公開日 | 2021-04-28 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTA
 
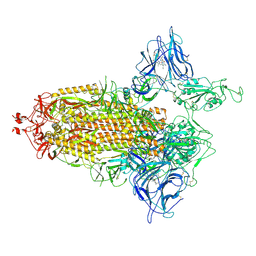 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | 登録日 | 2021-03-09 | | 公開日 | 2021-04-28 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTC
 
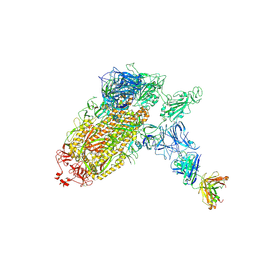 | | Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | 登録日 | 2021-03-09 | | 公開日 | 2021-04-28 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
6YA7
 
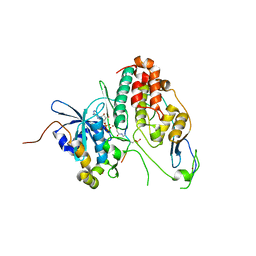 | | Cdc7-Dbf4 bound to an Mcm2-S40 derived bivalent substrate | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase, DNA replication licensing factor MCM2, ... | | 著者 | Dick, S.D, Cherepanov, P. | | 登録日 | 2020-03-11 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
6YA6
 
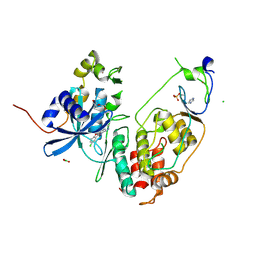 | | Minimal construct of Cdc7-Dbf4 bound to XL413 | | 分子名称: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, BORIC ACID, CHLORIDE ION, ... | | 著者 | Dick, S.D, Cherepanov, P. | | 登録日 | 2020-03-11 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
6YA8
 
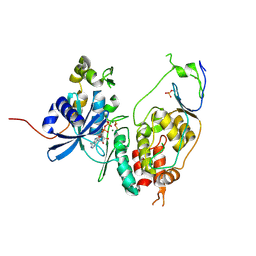 | | Cdc7-Dbf4 bound to ADP-BeF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase, ... | | 著者 | Dick, S.D, Cherepanov, P. | | 登録日 | 2020-03-11 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
7PEL
 
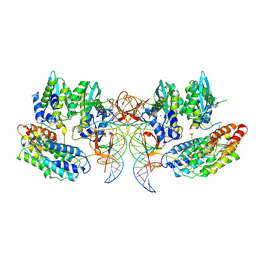 | | CryoEM structure of simian T-cell lymphotropic virus intasome in complex with PP2A regulatory subunit B56 gamma | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Isoform 3 of PC4 and SFRS1-interacting protein,Isoform Gamma-2 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | 著者 | Barski, M, Pye, V.E, Nans, A, Cherepanov, P, Maertens, G.N. | | 登録日 | 2021-08-10 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Cryo-EM structure of the deltaretroviral intasome in complex with the PP2A regulatory subunit B56gamma.
Nat Commun, 11, 2020
|
|
6R0C
 
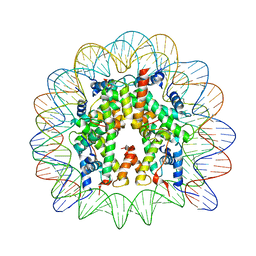 | | Human-D02 Nucleosome Core Particle with biotin-streptavidin label | | 分子名称: | DNA (142-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | 著者 | Pye, V.E, Wilson, M.D, Cherepanov, P, Costa, A. | | 登録日 | 2019-03-12 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | 分子名称: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | 著者 | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | 登録日 | 2019-05-09 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
8B7D
 
 | | Luminal domain of TMEM106B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | 著者 | Pye, V.E, Roustan, C, Cherepanov, P. | | 登録日 | 2022-09-29 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry.
Cell, 186, 2023
|
|
8A1P
 
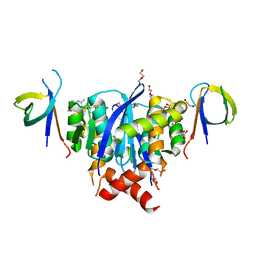 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BI-D | | 分子名称: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | 登録日 | 2022-06-01 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
8A1Q
 
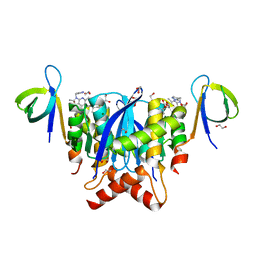 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor STP0404 (Pirmitegravir) | | 分子名称: | (2S)-tert-butoxy{4-(4-chlorophenyl)-2,3,6-trimethyl-1-[(1-methyl-1H-pyrazol-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridin-5-yl}acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | 著者 | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | 登録日 | 2022-06-01 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
8BUV
 
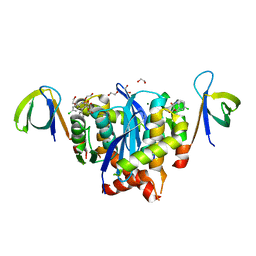 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor LEDGIN 3 | | 分子名称: | (6-chloro-2-oxo-4-phenyl-1,2-dihydroquinolin-3-yl)acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | 著者 | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | 登録日 | 2022-11-30 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor LEDGIN 3
To Be Published
|
|
6SP2
 
 | | CryoEM structure of SERINC from Drosophila melanogaster | | 分子名称: | CARDIOLIPIN, Lauryl Maltose Neopentyl Glycol, Membrane protein TMS1d, ... | | 著者 | Pye, V.E, Nans, A, Cherepanov, P. | | 登録日 | 2019-08-30 | | 公開日 | 2020-01-01 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | A bipartite structural organization defines the SERINC family of HIV-1 restriction factors.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8POE
 
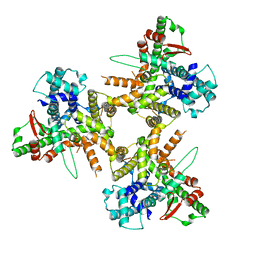 | | Structure of tissue-specific lipid scramblase ATG9B homotrimer, refined with C3 symmetry applied | | 分子名称: | Autophagy-related protein 9B | | 著者 | Chiduza, G.N, Pye, V.E, Tooze, S.A, Cherepanov, P. | | 登録日 | 2023-07-04 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | ATG9B is a tissue-specific homotrimeric lipid scramblase that can compensate for ATG9A.
Autophagy, 20, 2024
|
|
3OYK
 
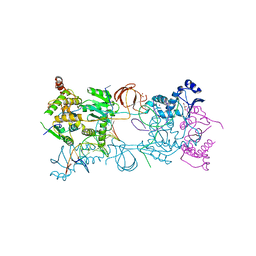 | | Crystal structure of the PFV S217H mutant intasome bound to manganese | | 分子名称: | AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | 著者 | Hare, S, Cherepanov, P. | | 登録日 | 2010-09-23 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYC
 
 | |
3OYN
 
 | | Crystal structure of the PFV N224H mutant intasome bound to magnesium and the INSTI MK2048 | | 分子名称: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | 著者 | Hare, S, Cherepanov, P. | | 登録日 | 2010-09-23 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYJ
 
 | | Crystal structure of the PFV S217Q mutant intasome in complex with magnesium and the INSTI MK2048 | | 分子名称: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | 著者 | Hare, S, Cherepanov, P. | | 登録日 | 2010-09-23 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYD
 
 | |
