4RA2
 
 | | PP2Ca | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2Ca
SCI REP, 2015
|
|
7MSL
 
 | |
6WCE
 
 | | Structure of the periplasmic binding protein P5PA | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal-5-phosphate binding protein A (P5PA) | | Authors: | Pan, C, Zimmer, A, Shah, M, Huynh, M, Lai, C.C.L, Sit, B, Hooda, Y, Moraes, T.F. | | Deposit date: | 2020-03-30 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Actinobacillus utilizes a binding protein-dependent ABC transporter to acquire the active form of vitamin B 6 .
J.Biol.Chem., 297, 2021
|
|
5Z9O
 
 | |
8GM3
 
 | | Vibrio harveyi Holo HphA | | Descriptor: | HEME B/C, Hemophilin | | Authors: | Pan, C, Shah, M, Moraes, T.F. | | Deposit date: | 2023-03-24 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Prevalence of Slam-dependent hemophilins in Gram-negative bacteria.
J.Bacteriol., 206, 2024
|
|
4RAG
 
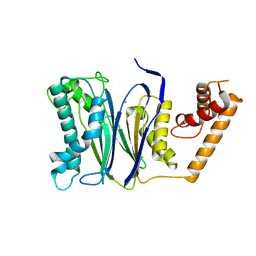 | | CRYSTAL STRUCTURE of PPC2A-D38K | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
4RAF
 
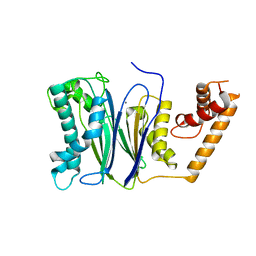 | | Crystal structure of PP2Ca-D38A | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
7UV9
 
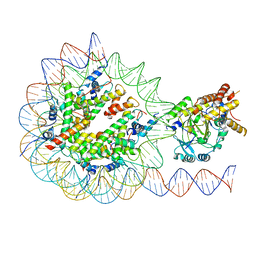 | | KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate | | Descriptor: | DNA (185-MER), FE (III) ION, Histone H2A type 1, ... | | Authors: | Spangler, C.J, Skrajna, A, Foley, C.A, Budziszewski, G.R, Azzam, D.N, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
2X1A
 
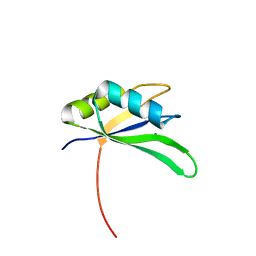 | | Structure of Rna15 RRM with RNA bound (G) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MAGNESIUM ION, MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1B
 
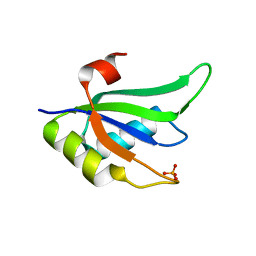 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
7TAN
 
 | | Structure of VRK1 C-terminal tail bound to nucleosome core particle | | Descriptor: | Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, Histone H3.2, ... | | Authors: | Spangler, C.J, Budziszewski, G.R, McGinty, R.K. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity.
Nucleic Acids Res., 50, 2022
|
|
3FY6
 
 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS3 | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
3G1J
 
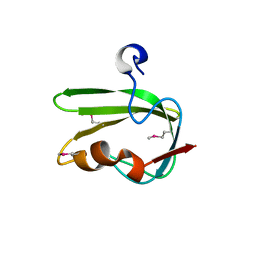 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4. | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4.
To be published
|
|
3W5J
 
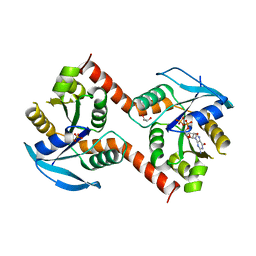 | | Crystal structure of GDP-bound NfeoB from Gallionella capsiferriformans | | Descriptor: | Ferrous iron transport protein B, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Deshpande, C.N, McGrath, A.P, Jormakka, M. | | Deposit date: | 2013-01-30 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure of an atypical FeoB G-domain reveals a putative domain-swapped dimer.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3GK6
 
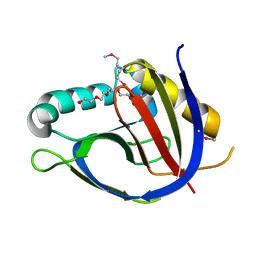 | | Crystal structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS2. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Integron cassette protein VCH_CASS2 | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Integron Gene Cassette-Associated Protein from Vibrio cholerae Identifies a Cationic Drug-Binding Module.
Plos One, 6, 2011
|
|
3I9S
 
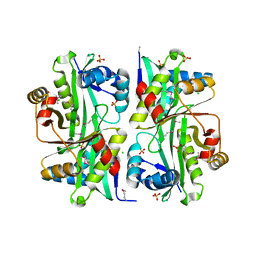 | | Structure from the mobile metagenome of V.cholerae. Integron cassette protein VCH_CASS6 | | Descriptor: | CHLORIDE ION, Integron cassette protein, SULFATE ION | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-13 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure from the mobile metagenome of V.cholerae. Integron cassette protein VCH_CASS6
To be Published
|
|
3IMO
 
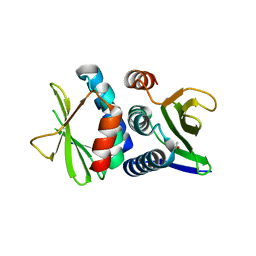 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS14 | | Descriptor: | ACETATE ION, Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites
Plos One, 8, 2013
|
|
3W5I
 
 | |
2X1F
 
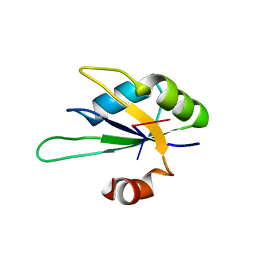 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
3KEH
 
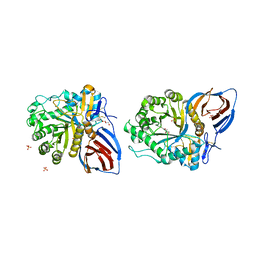 | | Crystal Structure of N370S Glucocerebrosidase mutant at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, R.R, Boucher, S, Pan, C.Q, Edmunds, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glucocerebrosidase Containing the N370S mutation: Implication on Chaperon Therapy
To be Published
|
|
3KE0
 
 | | Crystal structure of N370S Glucocerebrosidase at acidic pH. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, R.R, Boucher, S, Pan, C.Q, Edmunds, T. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Glucocerebrosidase Containing the N370S Mutation, Implications for Chaperon Therapy
To be Published
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
