3OB6
 
 | | Structure of AdiC(N101A) in the open-to-out Arg+ bound conformation | | Descriptor: | ARGININE, AdiC arginine:agmatine antiporter | | Authors: | Carpena, X, Kowalczyk, L, Ratera, M, Valencia, E, Vazquez-lbar, J.L, Fita, I, Palacin, M. | | Deposit date: | 2010-08-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of substrate-induced permeation by an amino acid antiporter.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2DH3
 
 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ZINC ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
2DH2
 
 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ACETATE ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
1U3C
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U3D
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2M1M
 
 | | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response | | Descriptor: | Auxin-induced protein IAA4 | | Authors: | Kovermann, M, Dinesh, D.C, Gopalswamy, M, Abel, S, Balbach, J. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1I7N
 
 | |
1DQ8
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG AND COA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1SIW
 
 | | Crystal structure of the apomolybdo-NarGHI | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rothery, R.A, Bertero, M.G, Cammack, R, Palak, M, Blasco, F, Strynadka, N.C, Weiner, J.H. | | Deposit date: | 2004-03-01 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic subunit of Escherichia coli nitrate reductase A contains a novel [4Fe-4S] cluster with a high-spin ground state
Biochemistry, 43, 2004
|
|
1DQ9
 
 | | COMPLEX OF CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG-COA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, PROTEIN (HMG-COA REDUCTASE) | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1DQA
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG, COA, AND NADP+ | | Descriptor: | 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1I7L
 
 | |
4GLN
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D-RFX001, Vascular endothelial growth factor A | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GLU
 
 | | Crystal structure of the mirror image form of VEGF-A | | Descriptor: | ACETATE ION, D- Vascular endothelial growth factor-A, GLYCEROL, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GLS
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D- RFX001, D- Vascular endothelial growth factor-A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1Y4Z
 
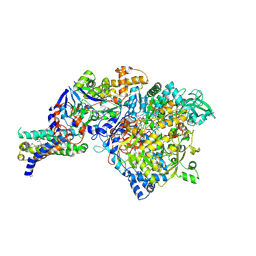 | | The crystal structure of Nitrate Reductase A, NarGHI, in complex with the Q-site inhibitor pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-01 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1FEP
 
 | | FERRIC ENTEROBACTIN RECEPTOR | | Descriptor: | FERRIC ENTEROBACTIN RECEPTOR | | Authors: | Buchanan, S.K, Smith, B.S, Ventatramani, L, Xia, D, Esser, L, Palnitkar, M, Chakraborty, R, Van Der Helm, D, Deisenhofer, J. | | Deposit date: | 1998-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the outer membrane active transporter FepA from Escherichia coli.
Nat.Struct.Biol., 6, 1999
|
|
1Y5L
 
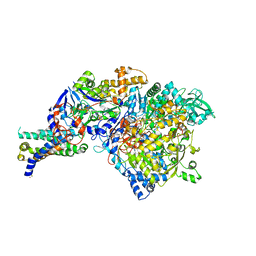 | | The crystal structure of the NarGHI mutant NarI-H66Y | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5N
 
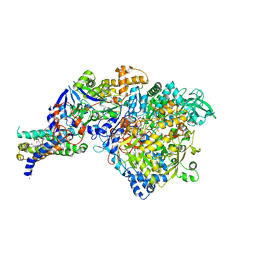 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5I
 
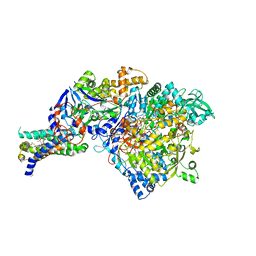 | | The crystal structure of the NarGHI mutant NarI-K86A | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
9H76
 
 | | Bacterial LAT transporter BASC in complex with L-Ala and NB53 | | Descriptor: | ALANINE, Nanobody NB53, Putative amino acid/polyamine transport protein | | Authors: | Fort, J, Palacin, M. | | Deposit date: | 2024-10-26 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The conserved lysine residue in transmembrane helix 5 is pivotal for the cytoplasmic gating of the L-amino acid transporters.
Pnas Nexus, 4, 2025
|
|
2V4X
 
 | | Crystal Structure of Jaagsiekte Sheep Retrovirus Capsid N-terminal domain | | Descriptor: | CAPSID PROTEIN P27 | | Authors: | Mortuza, G.B, Goldstone, D.C, Pashley, C, Haire, L.F, Palmarini, M, Taylor, W.R, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Capsid Amino-Terminal Domain from the Betaretrovirus, Jaagsiekte Sheep Retrovirus.
J.Mol.Biol., 386, 2009
|
|
2W1P
 
 | | 1.4 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 8.0 | | Descriptor: | AQUAPORIN PIP2-7 7;, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-20 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism
Plos Biol., 7, 2009
|
|
2W2E
 
 | | 1.15 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 3.5 | | Descriptor: | AQUAPORIN PIP2-7 7, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Plos Biol., 7, 2009
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
