4AZ4
 
 | |
4D5C
 
 | |
4ECA
 
 | |
4D5F
 
 | |
1QMI
 
 | | Crystal structure of RNA 3'-terminal phosphate cyclase, an ubiquitous enzyme with unusual topology | | Descriptor: | RNA 3'-TERMINAL PHOSPHATE CYCLASE | | Authors: | Palm, G.J, Billy, E, Filipowicz, W, Wlodawer, A. | | Deposit date: | 1999-09-28 | | Release date: | 2000-01-11 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of RNA 3'-Terminal Phosphate Cyclase, a Ubiquitous Enzyme with Unusual Topology
Structure, 8, 2000
|
|
2XQQ
 
 | | Human dynein light chain (DYNLL2) in complex with an in vitro evolved peptide (Ac-SRGTQTE). | | Descriptor: | ACETATE ION, DYNEIN LIGHT CHAIN 2, CYTOPLASMIC, ... | | Authors: | Rapali, P, Radnai, L, Suveges, D, Hetenyi, C, Harmat, V, Tolgyesi, F, Wahlgren, W.Y, Katona, G, Nyitray, L, Pal, G. | | Deposit date: | 2010-09-07 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Directed Evolution Reveals the Binding Motif Preference of the Lc8/Dynll Hub Protein and Predicts Large Numbers of Novel Binders in the Human Proteome
Plos One, 6, 2011
|
|
2PRK
 
 | |
1RN1
 
 | | THREE-DIMENSIONAL STRUCTURE OF GLN 25-RIBONUCLEASE T1 AT 1.84 ANGSTROMS RESOLUTION: STRUCTURAL VARIATIONS AT THE BASE RECOGNITION AND CATALYTIC SITES | | Descriptor: | RIBONUCLEASE T1 ISOZYME, SULFATE ION | | Authors: | Arni, R.K, Pal, G.P, Ravichandran, K.G, Tulinsky, A, Walz Junior, F.G, Metcalf, P. | | Deposit date: | 1991-11-22 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Three-dimensional structure of Gln25-ribonuclease T1 at 1.84-A resolution: structural variations at the base recognition and catalytic sites.
Biochemistry, 31, 1992
|
|
3P8M
 
 | | Human dynein light chain (DYNLL2) in complex with an in vitro evolved peptide dimerized by leucine zipper | | Descriptor: | Dynein light chain 2, General control protein GCN4 | | Authors: | Rapali, P, Radnai, L, Suveges, D, Hetenyi, C, Harmat, V, Tolgyesi, F, Wahlgren, W.Y, Katona, G, Nyitray, L, Pal, G. | | Deposit date: | 2010-10-14 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome.
Plos One, 6, 2011
|
|
2CTX
 
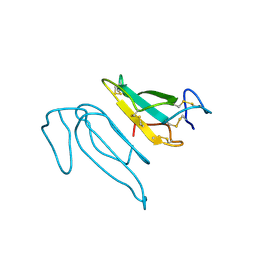 | | THE REFINED CRYSTAL STRUCTURE OF ALPHA-COBRATOXIN FROM NAJA NAJA SIAMENSIS AT 2.4-ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-COBRATOXIN | | Authors: | Betzel, C, Lange, G, Pal, G.P, Wilson, K.S, Maelicke, A, Saenger, W. | | Deposit date: | 1991-09-24 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The refined crystal structure of alpha-cobratoxin from Naja naja siamensis at 2.4-A resolution.
J.Biol.Chem., 266, 1991
|
|
2XTT
 
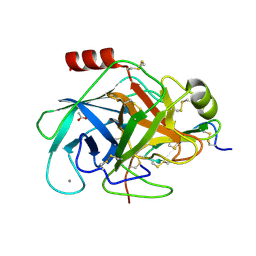 | | Bovine trypsin in complex with evolutionary enhanced Schistocerca gregaria protease inhibitor 1 (SGPI-1-P02) | | Descriptor: | ACETATE ION, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Wahlgren, W.Y, Pal, G, Kardos, J, Porrogi, P, Szenthe, B, Patthy, A, Graf, L, Katona, G. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The catalytic aspartate is protonated in the Michaelis complex formed between trypsin and an in vitro evolved substrate-like inhibitor: a refined mechanism of serine protease action.
J.Biol.Chem., 286, 2011
|
|
3TVJ
 
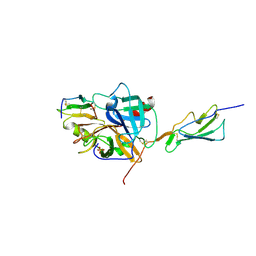 | | Catalytic fragment of MASP-2 in complex with its specific inhibitor developed by directed evolution on SGCI scaffold | | Descriptor: | Mannan-binding lectin serine protease 2 A chain, Mannan-binding lectin serine protease 2 B chain, Protease inhibitor SGPI-2, ... | | Authors: | Heja, D, Harmat, V, Dobo, J, Szasz, R, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2011-09-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
2OVI
 
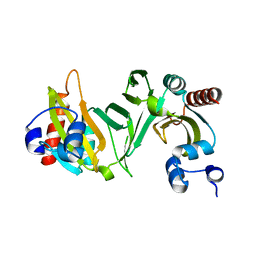 | |
4DJZ
 
 | | Catalytic fragment of masp-1 in complex with its specific inhibitor developed by directed evolution on sgci scaffold | | Descriptor: | Mannan-binding lectin serine protease 1 heavy chain, Mannan-binding lectin serine protease 1 light chain, Protease inhibitor SGPI-2 | | Authors: | Heja, D, Harmat, V, Fodor, K, Wilmanns, M, Dobo, J, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
1U5I
 
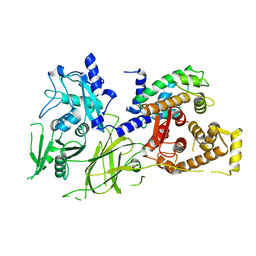 | | Crystal Structure analysis of rat m-calpain mutant Lys10 Thr | | Descriptor: | Calpain 2, large [catalytic] subunit precursor, Calpain small subunit 1 | | Authors: | Hosfield, C.M, Pal, G.P, Elce, J.S, Jia, Z. | | Deposit date: | 2004-07-27 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Activation of calpain by Ca2+: roles of the large subunit N-terminal and domain III-IV linker peptides
J.Mol.Biol., 343, 2004
|
|
7OB2
 
 | | NMR structure of the antimicrobial RiLK1 peptide in SDS micelles | | Descriptor: | RiLK1 | | Authors: | Falcigno, L, D'Auria, G, Palmieri, G, Gogliettino, M, Agrillo, B. | | Deposit date: | 2021-04-20 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Key Physicochemical Determinants in the Antimicrobial Peptide RiLK1 Promote Amphipathic Structures.
Int J Mol Sci, 22, 2021
|
|
4V08
 
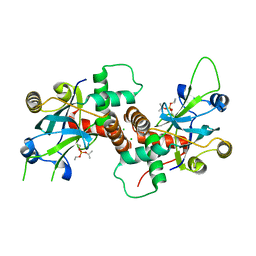 | | Inhibited dimeric pseudorabies virus protease pUL26N at 2 A resolution | | Descriptor: | CHLORIDE ION, DIISOPROPYL PHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
7Q5B
 
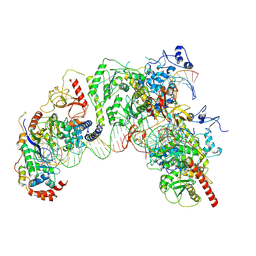 | | Cryo-EM structure of Ty3 retrotransposon targeting a TFIIIB-bound tRNA gene | | Descriptor: | DNA (19-MER), DNA (31-MER), DNA (34-MER), ... | | Authors: | Abascal-Palacios, G, Jochem, L, Pla-Prats, C, Beuron, F, Vannini, A. | | Deposit date: | 2021-11-03 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes.
Nat Commun, 12, 2021
|
|
2W3T
 
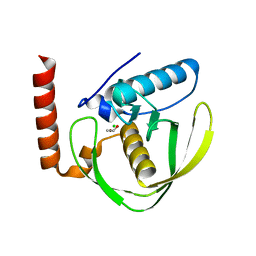 | | Chloro complex of the Ni-Form of E.coli deformylase | | Descriptor: | CHLORIDE ION, ETHANOL, NICKEL (II) ION, ... | | Authors: | Ngo, Y.H.T, Palm, G.J, Hinrichs, W. | | Deposit date: | 2008-11-14 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of the Ni(II) Complex of Escherichia Coli Peptide Deformylase and Suggestions on Deformylase Activities Depending on Different Metal(II) Centres.
J.Biol.Inorg.Chem., 15, 2010
|
|
7AST
 
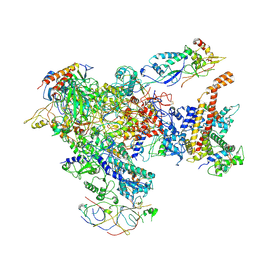 | | Apo Human RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Ramsay, E.P, Abascal-Palacios, G, Daiss, J.L, King, H, Gouge, J, Pilsl, M, Beuron, F, Morris, E, Gunkel, P, Engel, C, Vannini, A. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of human RNA polymerase III.
Nat Commun, 11, 2020
|
|
7ASV
 
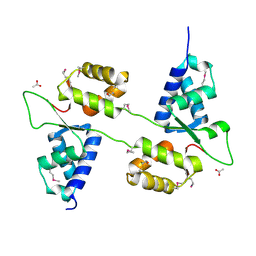 | |
5MIY
 
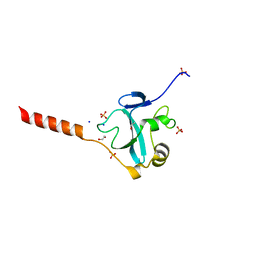 | | Crystal structure of the E3 ubiquitin ligase RavN from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin ligase RavN, SODIUM ION, ... | | Authors: | Lucas, M, Abascal-Palacios, G, Rojas, A.L, Hierro, A. | | Deposit date: | 2016-11-29 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | RavN is a member of a previously unrecognized group of Legionella pneumophila E3 ubiquitin ligases.
PLoS Pathog., 14, 2018
|
|
7ASU
 
 | |
5MWV
 
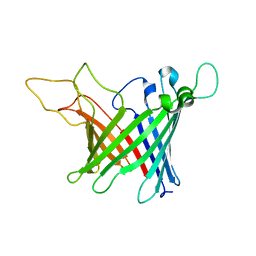 | | Solid-state NMR Structure of outer membrane protein G in lipid bilayers | | Descriptor: | Outer membrane protein G | | Authors: | Retel, J.S, Nieuwkoop, A.J, Hiller, M, Higman, V.A, Barbet-Massin, E, Stanek, J, Andreas, L.B, Franks, W.T, van Rossum, B.-J, Vinothkumar, K.R, Handel, L, de Palma, G.G, Bardiaux, B, Pintacuda, G, Emsley, L, Kuelbrandt, W, Oschkinat, H. | | Deposit date: | 2017-01-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of outer membrane protein G in lipid bilayers.
Nat Commun, 8, 2017
|
|
6EU3
 
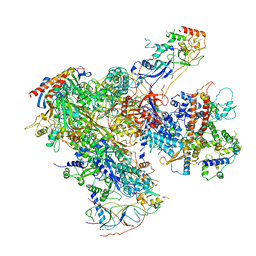 | | Apo RNA Polymerase III - closed conformation (cPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
