4GJF
 
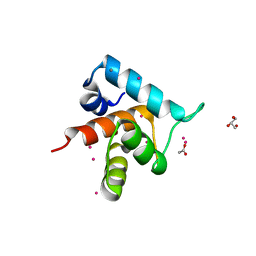 | |
4GJG
 
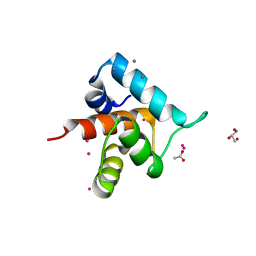 | |
1ERQ
 
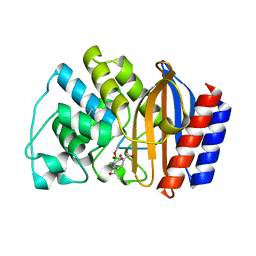 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-1-ACETAMIDO-2-(3-CARBOXY-2-HYDROXYPHENYL)ETHYL BORONIC ACID | | 分子名称: | 1(R)-1-ACETAMIDO-2-(3-CARBOXY-2-HYDROXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | 著者 | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | 登録日 | 2000-04-06 | | 公開日 | 2000-05-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
1ERO
 
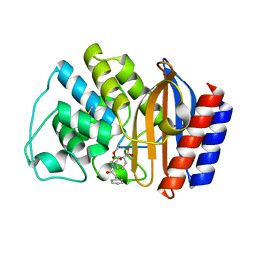 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-2-PHENYLACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID | | 分子名称: | (1R)-2-PHENYLACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | 著者 | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | 登録日 | 2000-04-06 | | 公開日 | 2000-05-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
2Q2I
 
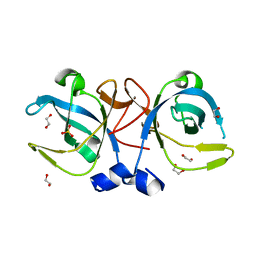 | | Crystal structure of the protein secretion chaperone CsaA from Agrobacterium tumefaciens. | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Secretion chaperone | | 著者 | Feldman, A.R, Shapova, Y.A, Paetzel, M. | | 登録日 | 2007-05-28 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Phage display and crystallographic analysis reveals potential substrate/binding site interactions in the protein secretion chaperone CsaA from Agrobacterium tumefaciens.
J.Mol.Biol., 379, 2008
|
|
3P06
 
 | |
2KXX
 
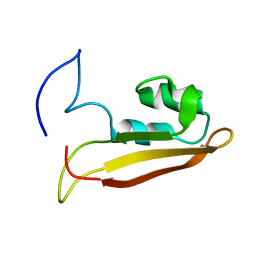 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | 分子名称: | Small protein A | | 著者 | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | 登録日 | 2010-05-13 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
3RV5
 
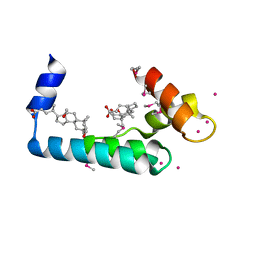 | | Crystal structure of human cardiac troponin C regulatory domain in complex with cadmium and deoxycholic acid | | 分子名称: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | 著者 | Li, A.Y, Lee, J, Borek, D, Otwinowski, Z, Tibbits, G, Paetzel, M. | | 登録日 | 2011-05-06 | | 公開日 | 2011-08-31 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of cardiac troponin C regulatory domain in complex with cadmium and deoxycholic Acid reveals novel conformation.
J.Mol.Biol., 413, 2011
|
|
2NZO
 
 | |
3SNS
 
 | | Crystal structure of the C-terminal domain of Escherichia coli lipoprotein BamC | | 分子名称: | CHLORIDE ION, Lipoprotein 34 | | 著者 | Kim, K.H, Aulakh, S, Tan, W, Paetzel, M. | | 登録日 | 2011-06-29 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystallographic analysis of the C-terminal domain of the Escherichia coli lipoprotein BamC.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3SD6
 
 | |
3RST
 
 | |
2MQ3
 
 | | NMR structure of the c3 domain of human cardiac myosin binding protein-c with a hypertrophic cardiomyopathy-related mutation R502W. | | 分子名称: | Myosin-binding protein C, cardiac-type | | 著者 | Zhang, X, De, S, Mcintosh, L.P, Paetzel, M. | | 登録日 | 2014-06-12 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of the C3 Domain of Cardiac Myosin Binding Protein C and Its Hypertrophic Cardiomyopathy-Related R502W Mutant.
Biochemistry, 53, 2014
|
|
2MQ0
 
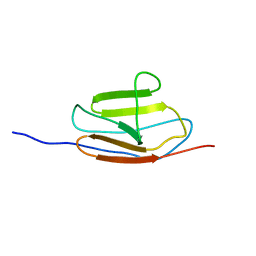 | | NMR structure of the c3 domain of human cardiac myosin binding protein-c | | 分子名称: | Myosin-binding protein C, cardiac-type | | 著者 | Zhang, X, De, S, Mcintosh, L.P, Paetzel, M. | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of the C3 Domain of Cardiac Myosin Binding Protein C and Its Hypertrophic Cardiomyopathy-Related R502W Mutant.
Biochemistry, 53, 2014
|
|
2NZH
 
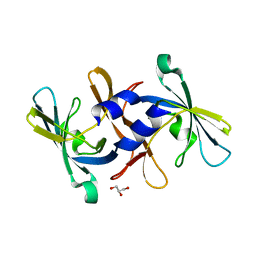 | |
3SWB
 
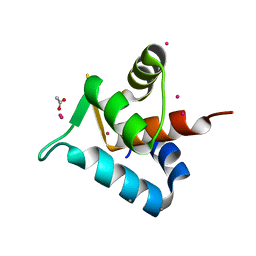 | |
3P1L
 
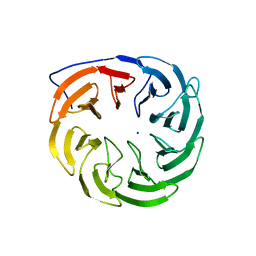 | | Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex, native crystals. | | 分子名称: | Lipoprotein yfgL, SODIUM ION | | 著者 | Kim, K.H, Paetzel, M. | | 登録日 | 2010-09-30 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex, native crystals
J.Mol.Biol., 406, 2011
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRY
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp12-nsp13 (C12) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp12-nsp13 (C12) cut site | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS0
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp14-nsp15 (C14) cut site sequence (form 2) | | 分子名称: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRZ
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp13-nsp14 (C13) cut site sequence | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS1
 
 | | Structure of SARS-CoV-2 Mpro in complex with nsp12-nsp13 (C12) cut site sequence | | 分子名称: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | 分子名称: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
