4GKI
 
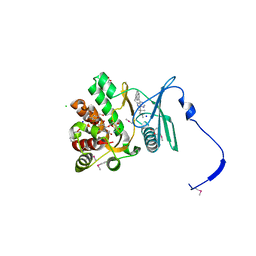 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NM-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4QL5
 
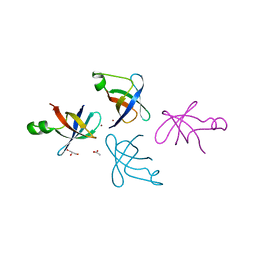 | | Crystal structure of translation initiation factor IF-1 from Streptococcus pneumoniae TIGR4 | | Descriptor: | ACETATE ION, GLYCEROL, Translation initiation factor IF-1, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Crystal structure of translation initiation factor IF-1 from Streptococcus pneumoniae TIGR4
TO BE PUBLISHED
|
|
4QOK
 
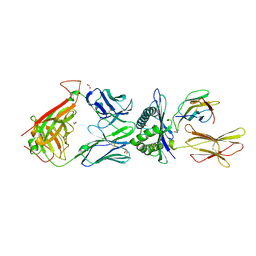 | | Structural basis for ineffective T-cell responses to MHC anchor residue improved heteroclitic peptides | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Madura, F, Sewell, A.K. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for ineffective T-cell responses to MHC anchor residue-improved "heteroclitic" peptides.
Eur.J.Immunol., 45, 2015
|
|
1NL9
 
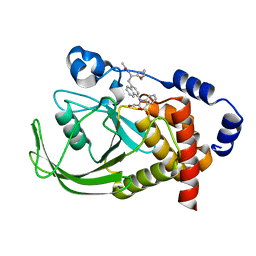 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 12 Using a Linked-Fragment Strategy | | Descriptor: | 2-{[4-(2-ACETYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-NAPHTHALEN-1-YL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine
Phosphatase 1B Inhibitor Using a
Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
4ILC
 
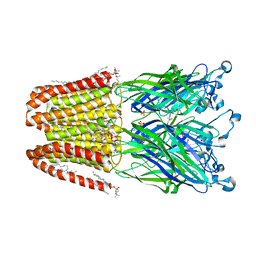 | | The GLIC pentameric ligand-gated ion channel in complex with sulfates | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
4RO3
 
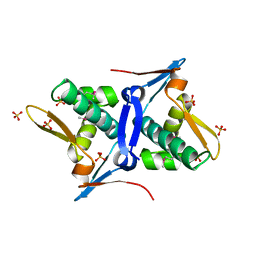 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | Hypothetical Protein, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-27 | | Release date: | 2014-12-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae.
To be Published
|
|
1QSG
 
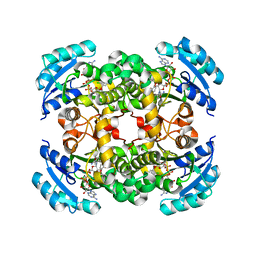 | | CRYSTAL STRUCTURE OF ENOYL REDUCTASE INHIBITION BY TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN, ... | | Authors: | Stewart, M.J, Parikh, S, Xiao, G, Tonge, P.J, Kisker, C. | | Deposit date: | 1999-06-21 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis and mechanism of enoyl reductase inhibition by triclosan.
J.Mol.Biol., 290, 1999
|
|
1Q8R
 
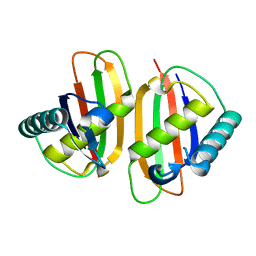 | | Structure of E.coli RusA Holliday junction resolvase | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Rafferty, J.B, Bolt, E.L, Muranova, T.A, Sedelnikova, S.E, Leonard, P, Pasquo, A, Baker, P.J, Rice, D.W, Sharples, G.J, Lloyd, R.G. | | Deposit date: | 2003-08-22 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold
Structure, 11, 2003
|
|
4IA9
 
 | | Crystal structure of human WD REPEAT DOMAIN 5 in complex with 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Bolshan, Y, Getlik, M, Tempel, W, Kuznetsova, E, Wasney, G.A, Hajian, T, Poda, G, Nguyen, K.T, Schapira, M, Brown, P.J, Al-awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Smil, D, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction.
ACS Med Chem Lett, 4, 2013
|
|
4I3F
 
 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
1Q89
 
 | |
4QNP
 
 | | Crystal structure of the 2009 pandemic H1N1 influenza virus neuraminidase with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wan, H.Q, Yang, H, Shore, D.A, Garten, R.J, Couzens, L, Gao, J, Jiang, L.L, Carney, P.J, Villanueva, J, Stevens, J, Eichelberger, M.C. | | Deposit date: | 2014-06-18 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers.
Nat Commun, 6, 2015
|
|
4HUR
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with acetyl coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
4QL1
 
 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
4I4W
 
 | | Peptide length determines the outcome of T cell receptor/peptide-MHCI engagement | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Wooldridge, L, Cole, D.K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-02 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide length determines the outcome of TCR/peptide-MHCI engagement.
Blood, 121, 2013
|
|
4QNK
 
 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4JCC
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiuA from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Iron-compound ABC transporter, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.133 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiuA from Streptococcus pneumoniae Canada MDR_19A
TO BE PUBLISHED
|
|
4JDV
 
 | | Crystal structure of germ-line precursor of NIH45-46 Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Diskin, R, Scharf, L, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1NZ7
 
 | | POTENT, SELECTIVE INHIBITORS OF PROTEIN TYROSINE PHOSPHATASE 1B USING A SECOND PHOSPHOTYROSINE BINDING SITE, complexed with compound 19. | | Descriptor: | 2-[(4-{2-ACETYLAMINO-2-[4-(1-CARBOXY-3-METHYLSULFANYL-PROPYLCARBAMOYL)-BUTYLCARBAMOYL]-ETHYL}-2-ETHYL-PHENYL)-OXALYL-AM INO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Xin, Z, Oost, T.K, Abad-Zapatero, C, Hajduk, P.J, Pei, Z, Szczepankiewicz, B.G, Hutchins, C.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Trevillyan, J.M, Jirousek, M.R, Liu, G. | | Deposit date: | 2003-02-16 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent, Selective Inhibitors of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
4IW7
 
 | | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis. | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Newcomb, W, Niedzialkowska, E, Porebski, P.J, Grimshaw, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis.
To be Published
|
|
4IT2
 
 | | Mn(III)-PPIX bound Tt H-NOX | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, Methyl-accepting chemotaxis protein | | Authors: | Winter, M.B, Klemm, P.J, Phillips-Piro, C.M, Raymond, K.M, Marletta, M.A. | | Deposit date: | 2013-01-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Porphyrin-Substituted H-NOX Proteins as High-Relaxivity MRI Contrast Agents.
Inorg.Chem., 52, 2013
|
|
1Q88
 
 | |
4IV9
 
 | | Structure of the Flavoprotein Tryptophan-2-Monooxygenase | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-INDOL-3-YL)ACETAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gaweska, H.M, Taylor, A.B, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structure of the flavoprotein tryptophan 2-monooxygenase, a key enzyme in the formation of galls in plants.
Biochemistry, 52, 2013
|
|
1Q87
 
 | |
