6MF0
 
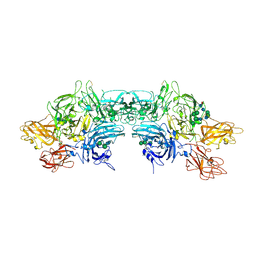 | | Crystal Structure Determination of Human/Porcine Chimera Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19999886 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
3I5K
 
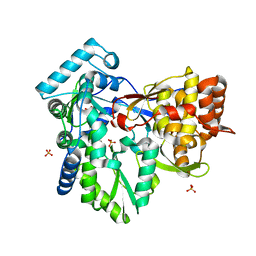 | | Crystal structure of the NS5B polymerase from Hepatitis C Virus (HCV) strain JFH1 | | Descriptor: | PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Simister, P.C, Schmitt, M, Lohmann, V, Bressanelli, S. | | Deposit date: | 2009-07-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of hepatitis C virus strain JFH1 polymerase
J.Virol., 83, 2009
|
|
5KT8
 
 | |
3H42
 
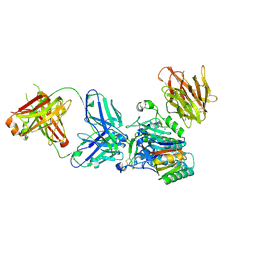 | | Crystal structure of PCSK9 in complex with Fab from LDLR competitive antibody | | Descriptor: | Fab from LDLR competitive antibody: Heavy chain, Fab from LDLR competitive antibody: Light chain, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T, Tsai, M.M, Yang, E. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H82
 
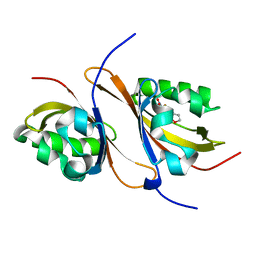 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS020 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(furan-2-ylmethyl)-2-nitro-4-(trifluoromethyl)aniline | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
3H7W
 
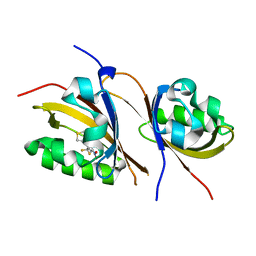 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS017 | | Descriptor: | 2-nitro-N-(thiophen-3-ylmethyl)-4-(trifluoromethyl)aniline, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
3HB9
 
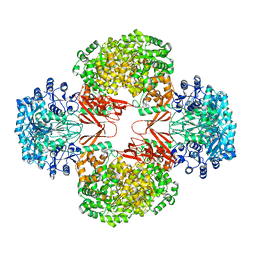 | | Crystal Structure of S. aureus Pyruvate Carboxylase A610T Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
1Z8V
 
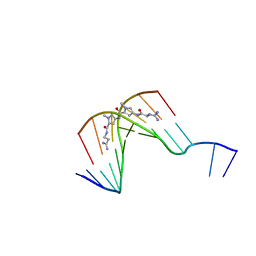 | | The Structure of d(GGCCAATTGG) Complexed with Netropsin | | Descriptor: | (5'-D(*GP*GP*CP*CP*AP*AP*TP*TP*GP*G)-3'), NETROPSIN | | Authors: | Van Hecke, K, Nam, P.C, Nguyen, M.T, Van Meervelt, L. | | Deposit date: | 2005-03-31 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Netropsin interactions in the minor groove of d(GGCCAATTGG) studied by a combination of resolution enhancement and ab initio calculations.
Febs J., 272, 2005
|
|
5KQ2
 
 | |
5KSN
 
 | |
5KQI
 
 | |
5KSG
 
 | |
3J7Y
 
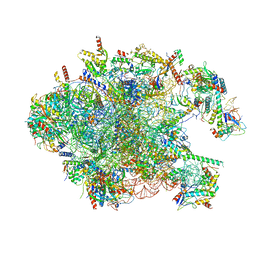 | | Structure of the large ribosomal subunit from human mitochondria | | Descriptor: | 16S rRNA, ADENOSINE MONOPHOSPHATE, CRIF1, ... | | Authors: | Brown, A, Amunts, A, Bai, X.C, Sugimoto, Y, Edwards, P.C, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the large ribosomal subunit from human mitochondria.
Science, 346, 2014
|
|
5KT9
 
 | |
5L02
 
 | | S324T variant of B. pseudomallei KatG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Catalase-peroxidase, PHOSPHATE ION, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the Ser324Thr variant of the catalase-peroxidase (KatG) from Burkholderia pseudomallei
J. Mol. Biol., 345, 2005
|
|
244D
 
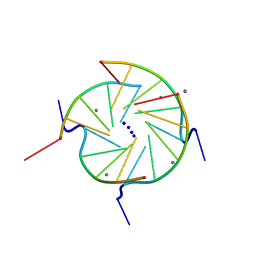 | | THE HIGH-RESOLUTION CRYSTAL STRUCTURE OF A PARALLEL-STRANDED GUANINE TETRAPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Laughlan, G, Murchie, A.I.H, Norman, D.G, Moore, M.H, Moody, P.C.E, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1995-10-19 | | Release date: | 1996-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The high-resolution crystal structure of a parallel-stranded guanine tetraplex.
Science, 265, 1994
|
|
1Y6R
 
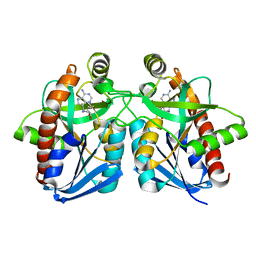 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1YN7
 
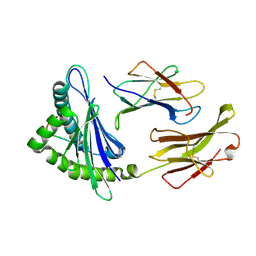 | | Crystal structure of a mouse MHC class I protein, H2-Db, in complex with a mutated peptide (R7A) of the influenza A acid polymerase | | Descriptor: | 10-mer peptide from RNA-directed RNA polymerase subunit P2, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Turner, S.J, Kedzierska, K, Komodromou, H, La Gruta, N.L, Dunstone, M.A, Webb, A.I, Webby, R, Walden, H, Xie, W, McCluskey, J, Purcell, A.W, Rossjohn, J, Doherty, P.C. | | Deposit date: | 2005-01-24 | | Release date: | 2005-06-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations
Nat.Immunol., 6, 2005
|
|
2A8C
 
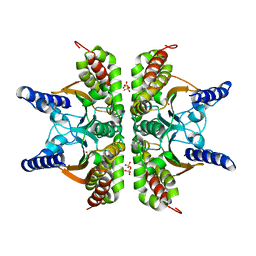 | | Haemophilus influenzae beta-carbonic anhydrase | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
2A8D
 
 | | Haemophilus influenzae beta-carbonic anhydrase complexed with bicarbonate | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
2A9E
 
 | |
2ABJ
 
 | | Crystal structure of human branched chain amino acid transaminase in a complex with an inhibitor, C16H10N2O4F3SCl, and pyridoxal 5' phosphate. | | Descriptor: | Branched-chain-amino-acid aminotransferase, cytosolic, N'-(5-CHLOROBENZOFURAN-2-CARBONYL)-2-(TRIFLUOROMETHYL)BENZENESULFONOHYDRAZIDE, ... | | Authors: | Ohren, J.F, Moreland, D.W, Rubin, J.R, Hu, H.L, McConnell, P.C, Mistry, A, Mueller, W.T, Scholten, J.D, Hasemann, C.H. | | Deposit date: | 2005-07-15 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The design and synthesis of human branched-chain amino acid aminotransferase inhibitors for treatment of neurodegenerative diseases.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2AEB
 
 | | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in immune response. | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2FZE
 
 | |
2GHD
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
