5ADV
 
 | | The Periplasmic Binding Protein CeuE of Campylobacter jejuni preferentially binds the iron(III) complex of the Linear Dimer Component of Enterobactin | | Descriptor: | 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, 2S-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-[(2S)-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-HYDROXY-PROPANOYL]OXY-PROPANOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Raines, D.J, Moroz, O.V, Turkenburg, J.P, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacteria in an Intense Competition for Iron: Key Component of the Campylobacter Jejuni Iron Uptake System Scavenges Enterobactin Hydrolysis Product.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1K9B
 
 | | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28 nm resolution. Structural peculiarities in a folded protein conformation | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Voss, R.H, Ermler, U, Essen, L.O, Wenzl, G, Kim, Y.M, Flecker, P. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28-nm resolution. Structural peculiarities in a folded protein conformation.
Eur.J.Biochem., 242, 1996
|
|
4BV0
 
 | | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Schuetz, M, Plueckthun, A. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1U1V
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | GLYCEROL, Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6S7W
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(quinolin-4-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
8EOZ
 
 | |
1YXP
 
 | | HIV-1 DIS RNA subtype F- Zn soaked | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', ZINC ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2005-02-22 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
2VG9
 
 | | Crystal structure of Loop Swap mutant of Necallimastix patriciarum Xyn11A | | Descriptor: | ACETATE ION, BIFUNCTIONAL ENDO-1,4-BETA-XYLANASE A, CADMIUM ION | | Authors: | Vardakou, M, Dumon, C, Flint, J.E, Murray, J.W, Christakopoulos, P, Weiner, D.P, Juge, N, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the Structural Basis for Substrate and Inhibitor Recognition in Eukaryotic Gh11 Xylanases.
J.Mol.Biol., 375, 2008
|
|
1Z5H
 
 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
1KBU
 
 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
3D0U
 
 | | Crystal Structure of Lysine Riboswitch Bound to Lysine | | Descriptor: | IRIDIUM HEXAMMINE ION, LYSINE, Lysine Riboswitch RNA | | Authors: | Garst, A.D, Heroux, A, Rambo, R.P, Batey, R.T. | | Deposit date: | 2008-05-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lysine riboswitch regulatory mRNA element.
J.Biol.Chem., 283, 2008
|
|
5NZ4
 
 | | Complex of I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Karlukova, E. | | Deposit date: | 2017-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
4GPX
 
 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-hydroxysisomicin (P212121 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
1YZ9
 
 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1O28
 
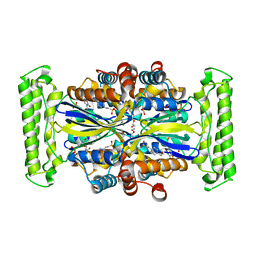 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FdUMP at 2.1 A resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, TRIETHYLENE GLYCOL, ... | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
2UZ9
 
 | | Human guanine deaminase (guaD) in complex with zinc and its product Xanthine. | | Descriptor: | GUANINE DEAMINASE, XANTHINE, ZINC ION | | Authors: | Moche, M, Welin, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human Guanine Deaminase (Guad) in Complex with Zinc and its Product Xhantine
To be Published
|
|
3D55
 
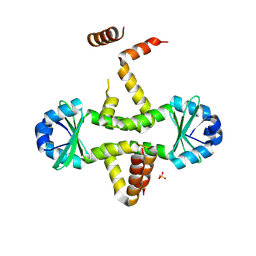 | | Crystal structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
5UDW
 
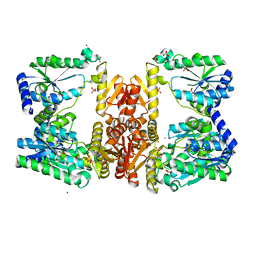 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with nickel | | Descriptor: | Lactate racemization operon protein LarE, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NUF
 
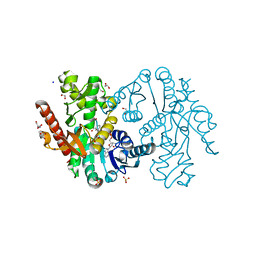 | | Cytosolic Malate Dehydrogenase 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
1ROC
 
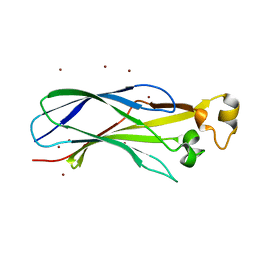 | | Crystal structure of the histone deposition protein Asf1 | | Descriptor: | Anti-silencing protein 1, BROMIDE ION | | Authors: | Daganzo, S.M, Erzberger, J.P, Lam, W.M, Skordalakes, E, Zhang, R, Franco, A.A, Brill, S.J, Adams, P.D, Berger, J.M, Kaufman, P.D. | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the conserved core of histone deposition protein Asf1.
Curr.Biol., 13, 2003
|
|
4H6C
 
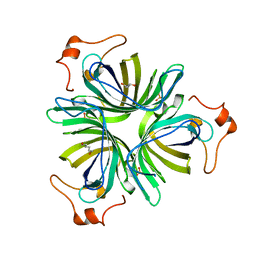 | |
5CFL
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p] | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CITRATE ANION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
7P9O
 
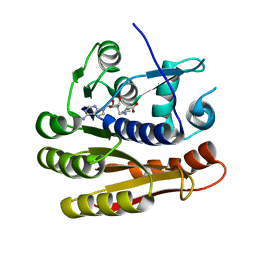 | | Structure of E.coli RlmJ in complex with a SAM analogue (CA) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]amino}-5'-deoxyadenosine, Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
2VGB
 
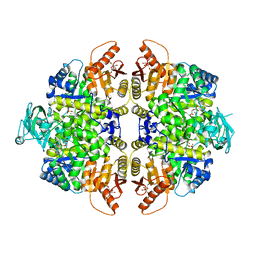 | | HUMAN ERYTHROCYTE PYRUVATE KINASE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
4C7X
 
 | | Thiamine Pyrophosphate Bound Transketolase from Lactobacillus salivarius at 2.2A resolution | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, TRANSKETOLASE | | Authors: | Lobley, C.M.C, Lukacik, P, Bumann, M, Aller, P, Douangamath, A, O'Toole, P.W, Walsh, M.A. | | Deposit date: | 2013-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | High Resolution Crystal Structures of Lactobacillus Salivarius Transketolase in the Presence and Absence of Thiamine Pyrophosphate
Acta Crystallogr.,Sect.F, 71, 2015
|
|
