4YAV
 
 | | Crystal structure of LigG in complex with B-glutathionyl-acetoveratrone (GS-AV) from Sphingobium sp. strain SYK-6 | | Descriptor: | Glutathione S-transferase, L-gamma-glutamyl-3-{(E)-[2-(3,4-dimethoxyphenyl)-2-oxoethylidene]-lambda~4~-sulfanyl}-L-alanylglycine, SULFATE ION | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
4YQN
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 6-{[(1R,3S,4R)-3-hydroxy-4-(hydroxymethyl)cyclopentyl]amino}pyridine-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
1PNL
 
 | |
6GSZ
 
 | | Crystal structure of native alfa-L-rhamnosidase from Aspergillus terreus | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Pachl, P, Rezacova, P, Skerlova, J. | | Deposit date: | 2018-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of native alpha-L-rhamnosidase from Aspergillus terreus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4YQO
 
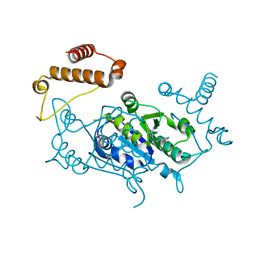 | |
4YTB
 
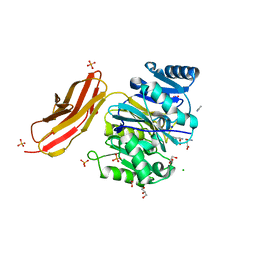 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in complex with dipeptide Asp-Gln. | | Descriptor: | ASPARTIC ACID, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
5N5Y
 
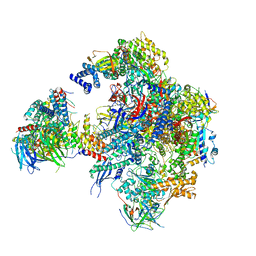 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation III) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
6RNJ
 
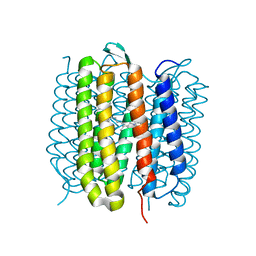 | | TR-SMX closed state structure (0-5ms) of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, F, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
8QJT
 
 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
4YQK
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 4-amino-N-(piperidin-4-yl)-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
6RZ8
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6RT4
 
 | |
6MVE
 
 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6NLF
 
 | | 1.45 A resolution structure of apo BfrB from Pseudomonas aeruginosa | | Descriptor: | Ferroxidase, POTASSIUM ION | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
4YVB
 
 | | Structure of D128N streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Baugh, L, Le Trong, I, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2015-03-19 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | A Streptavidin Binding Site Mutation Yields an Unexpected Result
To Be Published
|
|
8AJV
 
 | | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa crystallized in the presence of K+ cations | | Descriptor: | Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa crystallized in the presence of K+ cations
To Be Published
|
|
4Z3I
 
 | | Crystal structure of the lectin domain of PapG from E. coli BI47 in spacegroup P21212 | | Descriptor: | PapG, lectin domain | | Authors: | Jakob, R.P, Navarra, G, Zihlmann, P, Stangier, K, Preston, R.C, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | An unexpected way nature improves solvation entropy
To Be Published
|
|
8AJW
 
 | | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
6T7X
 
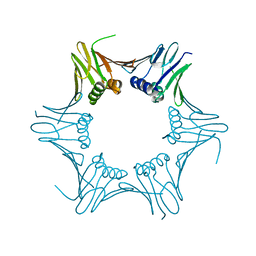 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
1D4L
 
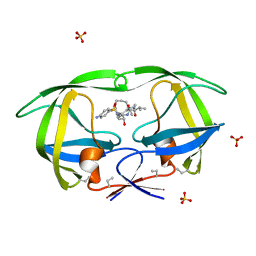 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
4Z3G
 
 | | Crystal structure of the lectin domain of PapG from E. coli BI47 in complex with 4-methoxyphenyl beta-D-galabiose in space group P212121 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methoxyphenol, PapG, ... | | Authors: | Jakob, R.P, Navarra, G, Zihlmann, P, Stangier, K, Preston, R.C, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An unexpected way nature improves solvation entropy
To Be Published
|
|
3K2B
 
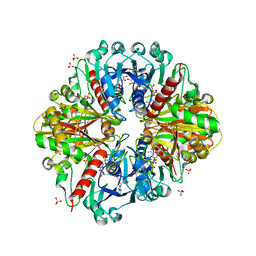 | | Crystal structure of photosynthetic A4 isoform glyceraldehyde-3-phosphate dehydrogenase complexed with NAD, from Arabidopsis thaliana | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Falini, G, Thumiger, A, Sparla, F, Marri, L, Trost, P. | | Deposit date: | 2009-09-29 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase (isoform A4) from Arabidopsis thaliana in complex with NAD
Acta Crystallogr.,Sect.F, 66, 2010
|
|
470D
 
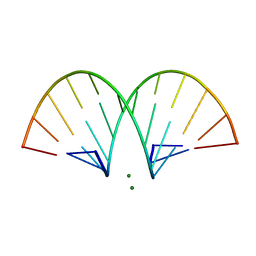 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
7U2B
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (53-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
6TUI
 
 | | Virion of empty GTA particle | | Descriptor: | Adaptor protein Rcc01688, Phage major capsid protein, HK97 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (10.47 Å) | | Cite: | Virion of empty GTA particle
Nat Commun, 11, 2020
|
|
