8DAH
 
 | | [20 bp edge] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.47 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8D4A
 
 | | Cas12a2 quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Nature, 613, 2023
|
|
6MJZ
 
 | | Cryo-EM structure of Human Parainfluenza Virus Type 3 (hPIV3) in complex with antibody PIA174 | | Descriptor: | Fusion glycoprotein F0, PIA174 Fab Heavy chain, PIA174 Fab Light chain | | Authors: | Acharya, P, Stewart-Jones, G, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4UC9
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal dipeptide of the Phosphoprotein | | Descriptor: | ASPARTIC ACID, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UC8
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal phenylalanine of the Phosphoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
8PIA
 
 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>T) | | Descriptor: | Chains: E, Chains: F, GLYCEROL, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
4UMW
 
 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN E2.PI STATE | | Descriptor: | MAGNESIUM ION, TETRAFLUOROALUMINATE ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
8PI8
 
 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA | | Descriptor: | Chains: E, Chains: F, GLYCEROL, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8PI9
 
 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>A) | | Descriptor: | Chains: E, Chains: F, Hepatocyte nuclear factor 1-alpha | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8PI7
 
 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -169C>T) | | Descriptor: | CHLORIDE ION, Chains: E, Chains: F, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8AMX
 
 | | AQP7 dimer of tetramers_D4 | | Descriptor: | Aquaporin-7 | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
8AMW
 
 | | AQP7 dimer of tetramers_C1 | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
6TAA
 
 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF ASPERGILLUS ORYZAE (TAKA) ALPHA-AMYLASE: AN APPLICATION OF THE SIMULATED-ANNEALING METHOD | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Swift, H.J, Brady, L, Derewenda, Z.S, Dodson, E.J, Turkenburg, J.P, Wilkinson, A.J. | | Deposit date: | 1992-08-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and molecular model refinement of Aspergillus oryzae (TAKA) alpha-amylase: an application of the simulated-annealing method.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
5ZYR
 
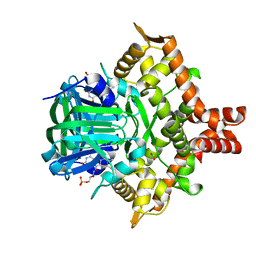 | | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, ... | | Authors: | Oonanant, W, Phongsak, T, Sucharitakul, J, Chaiyen, P, Yuvaniyama, J. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.20001316 Å) | | Cite: | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii
To Be Published
|
|
7GGO
 
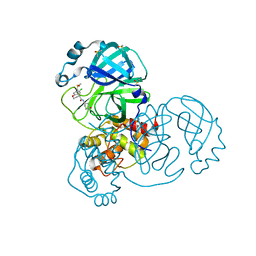 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-8b8a49e1-4 (Mpro-x12682) | | Descriptor: | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8FB6
 
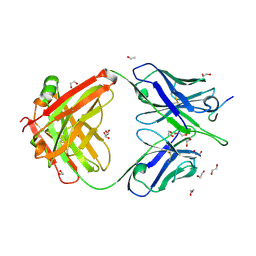 | | Crystal structure of Ky15.10 Fab in complex with circumsporozoite protein KQPA peptide | | Descriptor: | 1,2-ETHANEDIOL, Circumsporozoite protein KQPA peptide, Ky15.10 Antibody, ... | | Authors: | Prieto, K, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-29 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
7GGP
 
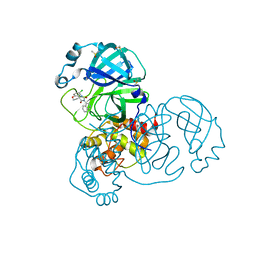 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with VLA-UCB-29506327-1 (Mpro-x12686) | | Descriptor: | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8FBA
 
 | |
7GH2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8AGU
 
 | | Yeast RQC complex in state E | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
7GH4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-00c1612e-1 (Mpro-x12777) | | Descriptor: | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8FDD
 
 | |
7GH5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-12 (Mpro-x2908) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8AKL
 
 | | Acyl-enzyme complex of meropenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2S,3R,4R)-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl-3-methyl-2-[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8F9U
 
 | |
