1K46
 
 | | Crystal Structure of the Type III Secretory Domain of Yersinia YopH Reveals a Domain-Swapped Dimer | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH | | Authors: | Smith, C.L, Khandelwal, P, Keliikuli, K, Zuiderweg, E.R.P, Saper, M.A. | | Deposit date: | 2001-10-05 | | Release date: | 2001-11-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the type III secretion and substrate-binding domain of Yersinia YopH phosphatase.
Mol.Microbiol., 42, 2001
|
|
5PYT
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 89) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PZ9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 105) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PZE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 110) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3K2J
 
 | | Crystal Structure of the 3rd Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | CHLORIDE ION, Protein polybromo-1, SULFATE ION | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Chaikuad, A, Pike, A.C.W, Krojer, T, Sethi, R, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the 3rd Bromodomain of Human Poly-bromodomain containing protein 1 (PB1)
To be Published
|
|
4CIH
 
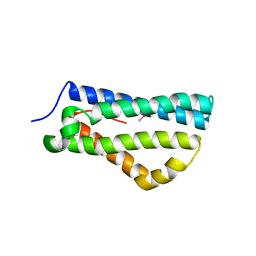 | | Structure of LntA-K180D-K181D from Listeria monocytogenes | | Descriptor: | LISTERIA NUCLEAR TARGETED PROTEIN A | | Authors: | Lebreton, A, Job, V, Ragon, M, Le Monnier, A, Dessen, A, Cossart, P, Bierne, H. | | Deposit date: | 2013-12-09 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA.
MBio, 5, 2014
|
|
1K4W
 
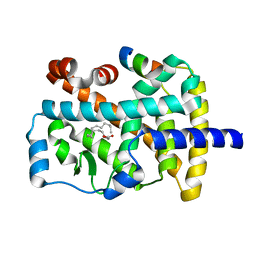 | | X-ray structure of the orphan nuclear receptor ROR beta ligand-binding domain in the active conformation | | Descriptor: | Nuclear receptor ROR-beta, STEARIC ACID, steroid receptor coactivator-1 | | Authors: | Stehlin, C, Wurtz, J.M, Steinmetz, A, Greiner, E, Schuele, R, Moras, D, Renaud, J.P. | | Deposit date: | 2001-10-09 | | Release date: | 2002-04-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation.
EMBO J., 20, 2001
|
|
3ERI
 
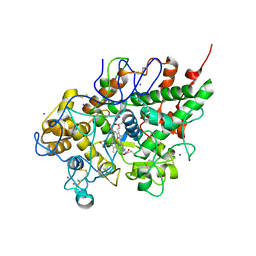 | | First structural evidence of substrate specificity in mammalian peroxidases: Crystal structures of substrate complexes with lactoperoxidases from two different species | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sheikh, I.A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-10-02 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
3NOV
 
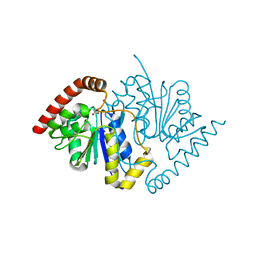 | | Crystal Structure of D17E Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
4CKB
 
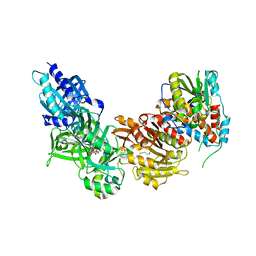 | | Vaccinia virus capping enzyme complexed with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, ... | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
7C5C
 
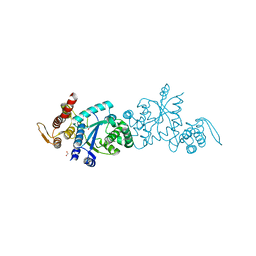 | |
2M08
 
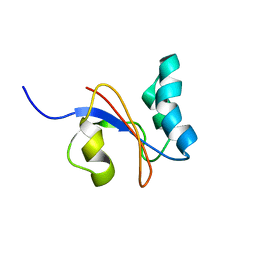 | |
3K43
 
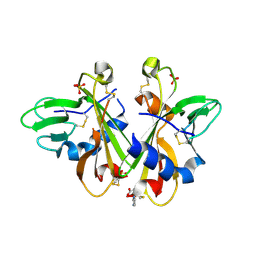 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
1BN0
 
 | | SL3 HAIRPIN FROM THE PACKAGING SIGNAL OF HIV-1, NMR, 11 STRUCTURES | | Descriptor: | SL3 RNA HAIRPIN | | Authors: | Pappalardo, L, Kerwood, D.J, Pelczer, I, Borer, P.N. | | Deposit date: | 1998-07-31 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional folding of an RNA hairpin required for packaging HIV-1.
J.Mol.Biol., 282, 1998
|
|
1BWX
 
 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-39, NMR, 10 STRUCTURES | | Descriptor: | PARATHYROID HORMONE | | Authors: | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | Deposit date: | 1998-09-29 | | Release date: | 2000-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
6AZX
 
 | | Crystal structure of the neutralizing anti-circumsporozoite protein 663 antibody | | Descriptor: | 663 antibody, heavy chain, light chain | | Authors: | Scally, S.W, Bosch, A, Triller, G, Wardemann, H, Julien, J.P. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies.
Immunity, 47, 2017
|
|
3RLP
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6B0C
 
 | | KLP10A-AMPPNP in complex with curved tubulin and a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
4OEK
 
 | | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYLETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kumar, M, Singh, R.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A
To be Published
|
|
2MDA
 
 | |
1TUQ
 
 | | NMR Structure Analysis of the B-DNA Dodecamer CTCtCACGTGGAG with a tricyclic cytosin base analogue | | Descriptor: | 5'-D(P*CP*TP*CP*(TC1)P*AP*CP*GP*TP*GP*GP*AP*G)-3' | | Authors: | Engman, K.C, Sandin, P, Osborne, S, Brown, T, Billeter, M, Lincoln, P, Norden, B, Albinsson, B, Wilhelmsson, L.M. | | Deposit date: | 2004-06-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-Vis spectroscopy characterization.
Nucleic Acids Res., 32, 2004
|
|
1BSK
 
 | | ZINC DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, PHOSPHATE ION, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
6IGR
 
 | | Crystal structure of S9 peptidase (S514A mutant in inactive state) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Gaur, N.K, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
3NTK
 
 | | Crystal structure of Tudor | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
5PQP
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 62) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
