1Q61
 
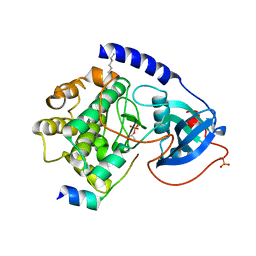 | | PKA triple mutant model of PKB | | Descriptor: | N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
3HTB
 
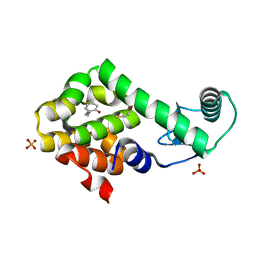 | | 2-propylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-propylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
1XCO
 
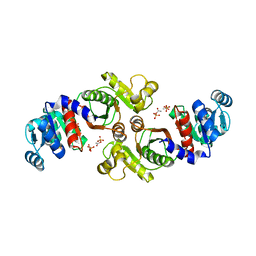 | | Crystal Structure of a Phosphotransacetylase from Bacillus subtilis in complex with acetylphosphate | | Descriptor: | ACETYLPHOSPHATE, Phosphate acetyltransferase, SULFATE ION | | Authors: | Xu, Q.S, Jancarik, J, Lou, Y, Yokota, H, Adams, P, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-09-02 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of a phosphotransacetylase from Bacillus subtilis and its complex with acetyl phosphate
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1XDK
 
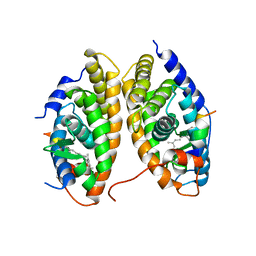 | | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator | | Descriptor: | (9cis)-retinoic acid, Retinoic acid receptor RXR-alpha, Retinoic acid receptor, ... | | Authors: | Pogenberg, V, Guichou, J.F, Vivat-Hannah, V, Kammerer, S, Perez, E, Germain, P, De Lera, A.R, Gronemeyer, H, Royer, C.A, Bourguet, W. | | Deposit date: | 2004-09-07 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CHARACTERIZATION OF THE INTERACTION BETWEEN RAR/RXR HETERODIMERS AND TRANSCRIPTIONAL COACTIVATORS THROUGH STRUCTURAL AND FLUORESCENCE ANISOTROPY STUDIES
J.Biol.Chem., 280, 2005
|
|
2K2U
 
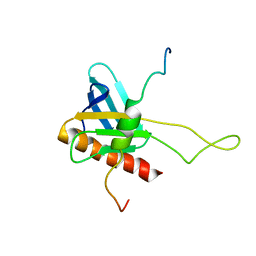 | | NMR Structure of the complex between Tfb1 subunit of TFIIH and the activation domain of VP16 | | Descriptor: | Alpha trans-inducing protein, RNA polymerase II transcription factor B subunit 1 | | Authors: | Langlois, C, Mas, C, Di Lello, P, Miller Jenkins, P.M, Legault, J, Omichinski, J.G. | | Deposit date: | 2008-04-11 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Complex between the Tfb1 Subunit of TFIIH and the Activation Domain of VP16: Structural Similarities between VP16 and p53.
J.Am.Chem.Soc., 130, 2008
|
|
1K45
 
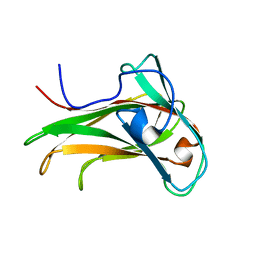 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
2ZO5
 
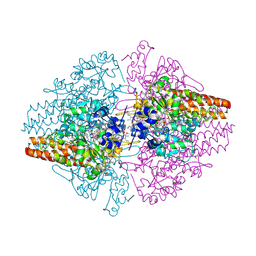 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with azide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
4PSG
 
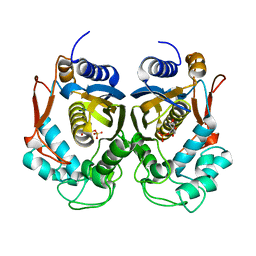 | |
2EHA
 
 | | Crystal structure of goat lactoperoxidase complexed with formate anion at 3.3 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Ethayathulla, A.S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of goat lactoperoxidase complexed with formate anion at 3.3 A resolution
to be published
|
|
4N2A
 
 | | Crystal structure of Protein Arginine Deiminase 2 (5 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2E
 
 | | Crystal structure of Protein Arginine Deiminase 2 (D123N, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4GXN
 
 | |
3HT7
 
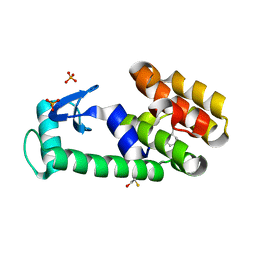 | | 2-ethylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTF
 
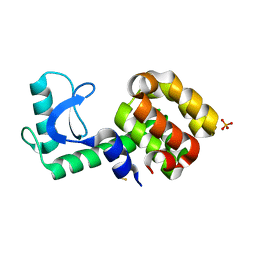 | | 4-chloro-1h-pyrazole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4-chloro-1H-pyrazole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
4GY5
 
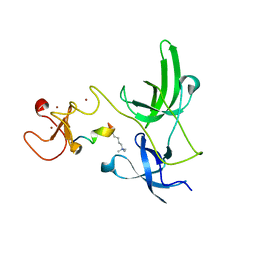 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
4REK
 
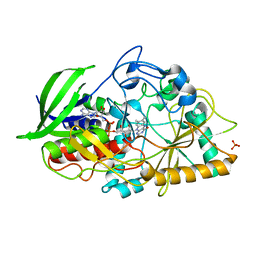 | | Crystal structure and charge density studies of cholesterol oxidase from Brevibacterium sterolicum at 0.74 ultra-high resolution | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zarychta, B, Lyubimov, A, Ahmed, M, Munshi, P, Guillot, B, Vrielink, A. | | Deposit date: | 2014-09-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Cholesterol oxidase: ultrahigh-resolution crystal structure and multipolar atom model-based analysis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5O7Y
 
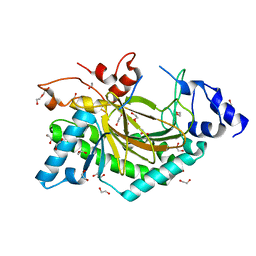 | | Thebaine 6-O-demethylase (T6ODM) from Papaver somniferum in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICKEL (II) ION, ... | | Authors: | Kluza, A, Niedzialkowska, E, Kurpiewska, K, Porebski, P.J, Borowski, T. | | Deposit date: | 2017-06-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of thebaine 6-O-demethylase from the morphine biosynthesis pathway.
J. Struct. Biol., 202, 2018
|
|
6ZYY
 
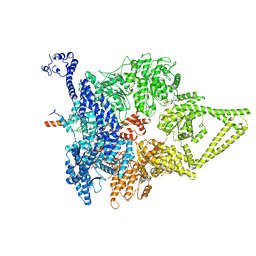 | | Outer Dynein Arm-Shulin complex - Dyh3 motor region (Tetrahymena thermophila) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Mali, G.R, Abid Ali, F, Lau, C.K, Begum, F, Boulanger, J, Howe, J.D, Chen, Z.A, Rappsilber, J, Skehel, M, Carter, A.P. | | Deposit date: | 2020-08-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Shulin packages axonemal outer dynein arms for ciliary targeting.
Science, 371, 2021
|
|
5OA0
 
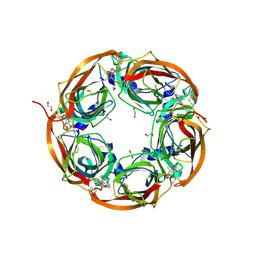 | | Crystal structure of mutant AChBP in complex with strychnine (T53F, Q74R, Y110A, I135S, W164F) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, STRYCHNINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
5OAN
 
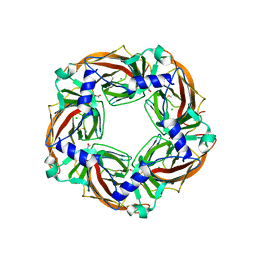 | | Crystal structure of mutant AChBP in complex with glycine (T53F, Q74R, Y110A, I135S, G162E, S206CCP_KGTG) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-23 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
4NB1
 
 | | Crystal Structure of FosB from Staphylococcus aureus at 1.80 Angstrom Resolution with L-Cysteine-Cys9 Disulfide | | Descriptor: | CYSTEINE, Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
2EF6
 
 | | Canavalia gladiata lectin complexed with Man1-3Man-OMe | | Descriptor: | CALCIUM ION, Concanavalin A, MANGANESE (II) ION, ... | | Authors: | Moreno, F.B.M.B, Bezerra, G.A, Oliveira, T.M, de Souza, E.P, da Rocha, B.A.M, Benevides, R.G, Delatorre, P, Cavada, B.S, de Azevedo Jr, W.F. | | Deposit date: | 2007-02-21 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: New insights into the understanding of the structure-biological activity relationship in legume lectins
J.Struct.Biol., 160, 2007
|
|
7AAY
 
 | | Crystal structure of MerTK kinase domain in complex with Merestinib | | Descriptor: | CHLORIDE ION, N-(3-fluoro-4-{[1-methyl-6-(1H-pyrazol-4-yl)-1H-indazol-5-yl]oxy}phenyl)-1-(4-fluorophenyl)-6-methyl-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Pflug, A, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
4ENQ
 
 | | Structure of E530D variant E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of main channel structure on H(2)O(2) access to the heme cavity of catalase KatE of Escherichia coli.
Arch.Biochem.Biophys., 526, 2012
|
|
7AB2
 
 | | Crystal structure of MerTK kinase domain in complex with UNC2025 | | Descriptor: | 4-[2-(butylamino)-5-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexan-1-ol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
