2V6U
 
 | |
5PK4
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 89) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3CQQ
 
 | | Human SOD1 G85R Variant, Structure II | | Descriptor: | ACETYL GROUP, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Cao, X, Antonyuk, S, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
5PKH
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 102) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1KHE
 
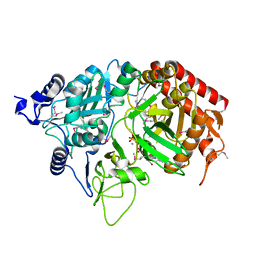 | | PEPCK complex with nonhydrolyzable GTP analog, MAD data | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Phosphoenolpyruvate Carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
2VN9
 
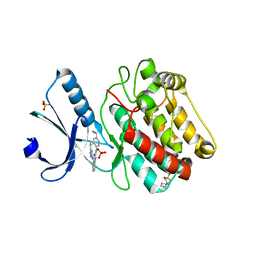 | | Crystal Structure of Human Calcium Calmodulin dependent Protein Kinase II delta isoform 1, CAMKD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II DELTA CHAIN, CHLORIDE ION, ... | | Authors: | Roos, A.K, Rellos, P, Salah, E, Pike, A.C.W, Fedorov, O, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Camkiidelta/Calmodulin Complex Reveals the Molecular Mechanism of Camkii Kinase Activation.
Plos Biol., 8, 2010
|
|
1KKP
 
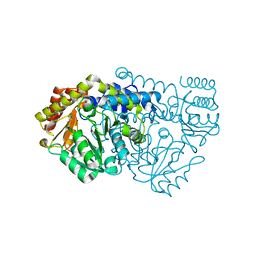 | | Crystal Structure of Serine Hydroxymethyltransferase complexed with Serine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
3N5N
 
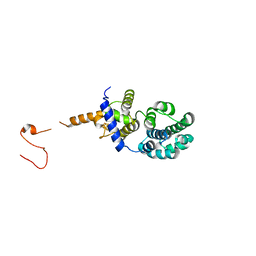 | |
2OJ0
 
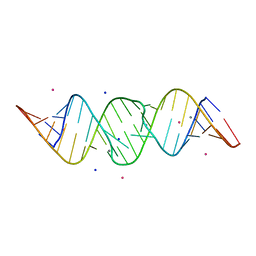 | | Crystal structure of the duplex form of the HIV-1(LAI) RNA dimerization initiation site MN soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2007-01-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cation-dependent cleavage of the duplex form of the subtype-B HIV-1 RNA dimerization initiation site.
Nucleic Acids Res., 38, 2010
|
|
5EI2
 
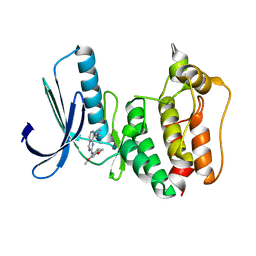 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-(2,4-dimethoxyphenyl)-8-(1-methylpyrazol-4-yl)pyrido[3,4-d]pyrimidin-2-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
5AEP
 
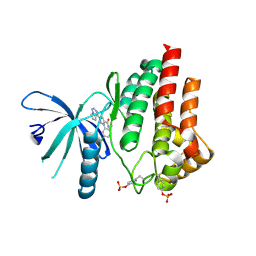 | | Novel pyrrole carboxamide inhibitors of JAK2 as potential treatment of myeloproliferative disorders | | Descriptor: | 1-(5-chloro-2-methylphenyl)-4-(pyrrolo[2,1-f][1,2,4]triazin-4-yl)-1H-pyrrole-2-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Bertrand, J, Brasca, M.G, Nesi, M, Amboldi, N, Avanzi, N, Bindi, S, Casero, D, Ciomei, M, Colombo, N, Cribioli, S, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Motto, I, Panzeri, A, Gnocchi, P, Donati, D. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Pyrrole Carboxamide Inhibitors of Jak2 as Potential Treatment of Myeloproliferative Disorders.
Bioorg.Med.Chem., 23, 2015
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
2VE7
 
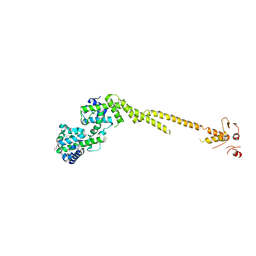 | | Crystal structure of a bonsai version of the human Ndc80 complex | | Descriptor: | GLYCEROL, KINETOCHORE PROTEIN HEC1, KINETOCHORE PROTEIN SPC25, ... | | Authors: | Ciferri, C, Pasqualato, S, Dos Reis, G, Screpanti, E, Maiolica, A, Polka, J, De Luca, J.G, De Wulf, P, Salek, M, Rappsilber, J, Moores, C.A, Salmon, E.D, Musacchio, A. | | Deposit date: | 2007-10-17 | | Release date: | 2008-05-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Implications for Kinetochore-Microtubule Attachment from the Structure of an Engineered Ndc80 Complex
Cell(Cambridge,Mass.), 133, 2008
|
|
5EP0
 
 | | Quorum-Sensing Signal Integrator LuxO - Receiver+Catalytic Domains | | Descriptor: | 1,2-ETHANEDIOL, Putative repressor protein luxO, SULFATE ION | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
3N7C
 
 | | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii | | Descriptor: | ABR034Wp | | Authors: | Sampathkumar, P, Manglicmot, D, Gilmore, J, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii
To be Published
|
|
5AFB
 
 | | Crystal structure of the Latrophilin3 Lectin and Olfactomedin Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Jackson, V.A, del Toro, D, Carrasquero, M, Roversi, P, Harlos, K, Klein, R, Seiradake, E. | | Deposit date: | 2015-01-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis of Latrophilin-Flrt Interaction.
Structure, 23, 2015
|
|
3CW6
 
 | | E. coli Initiator tRNA | | Descriptor: | Initiator tRNA | | Authors: | Barraud, P, Schmitt, E, Mechulam, Y, Dardel, F, Tisne, C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis.
Nucleic Acids Res., 36, 2008
|
|
5ADW
 
 | | The Periplasmic Binding Protein CeuE of Campylobacter jejuni preferentially binds the iron(III) complex of the Linear Dimer Component of Enterobactin | | Descriptor: | 2S-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-[(2S)-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-HYDROXY-PROPANOYL]OXY-PROPANOIC ACID, DIMETHYL SULFOXIDE, ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, ... | | Authors: | Raines, D.J, Moroz, O.V, Turkenburg, J.P, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacteria in an Intense Competition for Iron: Key Component of the Campylobacter Jejuni Iron Uptake System Scavenges Enterobactin Hydrolysis Product.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EPW
 
 | | C-Terminal Domain Of Human Coronavirus Nl63 Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Szelazek, B, Kabala, W, Kus, K, Zdzalik, M, Golik, P, Florek, D, Burmistrz, M, Pyrc, K, Dubin, G. | | Deposit date: | 2015-11-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
2VFZ
 
 | | CRYSTAL STRUCTURE OF ALPHA-1,3 GALACTOSYLTRANSFERASE (R365K) IN COMPLEX WITH UDP-2F-GALACTOSE | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYL TRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUOROGALACTOSE | | Authors: | Jamaluddin, H, Tumbale, P, Withers, S.G, Acharya, K.R, Brew, K. | | Deposit date: | 2007-11-06 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Binding Udp-2F-Galactose to Alpha-1,3 Galactosyltransferase-Implications for Catalysis
J.Mol.Biol., 369, 2007
|
|
5I9L
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT404 | | Descriptor: | 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
2VIX
 
 | | Methylated Shigella flexneri MxiC | | Descriptor: | ACETATE ION, GLYCEROL, PROTEIN MXIC | | Authors: | Deane, J.E, Roversi, P, King, C, Johnson, S, Lea, S.M. | | Deposit date: | 2007-12-05 | | Release date: | 2008-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of the Shigella Flexneri Type 3 Secretion System Protein Mxic Reveal Conformational Variability Amongst Homologues.
J.Mol.Biol., 377, 2008
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
2VK2
 
 | | Crystal structure of a galactofuranose binding protein | | Descriptor: | ABC TRANSPORTER PERIPLASMIC-BINDING PROTEIN YTFQ, beta-D-galactofuranose | | Authors: | Muller, A, Horler, R.S.P, Thomas, G.H, Wilson, K.S. | | Deposit date: | 2007-12-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Furanose-Specific Sugar Transport: Characterization of a Bacterial Galactofuranose-Binding Protein.
J.Biol.Chem., 284, 2009
|
|
2V9U
 
 | | Rim domain of main porin from Mycobacteria smegmatis | | Descriptor: | MSPA | | Authors: | Grueninger, D, Ziegler, M.O.P, Koetter, J.W.A, Treiber, N, Schulze, M.-S, Schulz, G.E. | | Deposit date: | 2007-08-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Designed Protein-Protein Association.
Science, 319, 2008
|
|
