4BNF
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2- phenoxy-5-propylphenol | | Descriptor: | 2-phenoxy-5-propyl-phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
6DW2
 
 | |
2HGF
 
 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
4OQQ
 
 | |
3GUY
 
 | | Crystal structure of a short-chain dehydrogenase/reductase from Vibrio parahaemolyticus | | Descriptor: | Short-chain dehydrogenase/reductase SDR | | Authors: | Patskovsky, Y, Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase from Vibrio parahaemolyticus
To be Published
|
|
6UE0
 
 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
2YIW
 
 | | triazolopyridine inhibitors of p38 kinase | | Descriptor: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthwaite, R.A, Mahnke, A, Mathais, J.P, Philip, J, Phillips, C, Smith, R.T, Stefamiak, M.H, Yeadon, M. | | Deposit date: | 2011-05-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
4BNM
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-hexyl- 2-(2-methylphenoxy)phenol | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
6L59
 
 | | Crystal structure of the alpha gamma heterodimer of human IDH3 in complex with CIT, Mg and ATP binding at allosteric site and Mg, ATP binding at active site. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Isocitrate dehydrogenase [NAD] subunit alpha, ... | | Authors: | Sun, P, Ding, J. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Molecular mechanism of the dual regulatory roles of ATP on the alpha gamma heterodimer of human NAD-dependent isocitrate dehydrogenase.
Sci Rep, 10, 2020
|
|
3H0A
 
 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Gamma (PPARg) and Retinoic Acid Receptor Alpha (RXRa) in Complex with 9-cis Retinoic Acid, Co-activator Peptide, and a Partial Agonist | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 1, Co-activator Peptide, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4KAP
 
 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | Descriptor: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4BNK
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-fluoro- 2-phenoxyphenol | | Descriptor: | 5-fluoro-2-phenoxyphenol, CHLORIDE ION, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
2BGW
 
 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
5ZRL
 
 | | M. smegmatis antimutator protein MutT2 in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-DIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
6IUG
 
 | | Cryo-EM structure of the plant actin filaments from Zea mays pollen | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, pollen F-actin | | Authors: | Ren, Z.H, Zhang, Y, Zhang, Y, He, Y.Q, Du, P.Z, Wang, Z.X, Sun, F, Ren, H.Y. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of Actin Filaments fromZea maysPollen.
Plant Cell, 31, 2019
|
|
5E89
 
 | | Crystal structure of Human galectin-3 CRD in complex with 3-fluophenyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
8BW9
 
 | | Cryo-EM structure of the RAF activating complex KSR-MEK-CNK-HYP | | Descriptor: | Connector enhancer of KSR protein CNK, Dual specificity mitogen-activated protein kinase kinase dSOR1, KSR, ... | | Authors: | Maisonneuve, P, Fronzes, R, Sicheri, F. | | Deposit date: | 2022-12-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The CNK-HYP scaffolding complex promotes RAF activation by enhancing KSR-MEK interaction.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5E8A
 
 | | Crystal structure of Human galectin-3 CRD in complex with 4-fluophenyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
3LKK
 
 | |
5EAP
 
 | | Crystal structure of human WDR5 in complex with compound 9e | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
6TVI
 
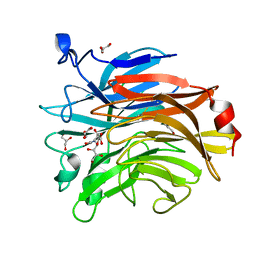 | | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA
To Be Published
|
|
8BVX
 
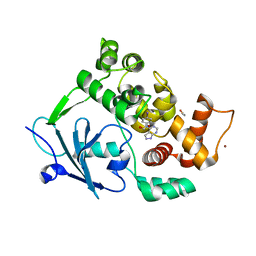 | |
5OOF
 
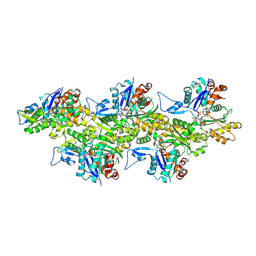 | | Cryo-EM structure of F-actin in complex with ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2017-08-07 | | Release date: | 2018-06-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5DTF
 
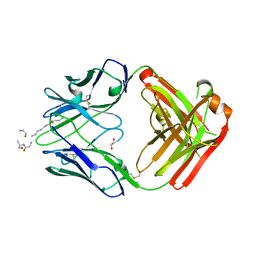 | |
2FAM
 
 | | X-RAY CRYSTAL STRUCTURE OF FERRIC APLYSIA LIMACINA MYOGLOBIN IN DIFFERENT LIGANDED STATES | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Conti, E, Moser, C, Rizzi, M, Mattevi, A, Lionetti, C, Coda, A, Ascenzi, P, Brunori, M, Bolognesi, M. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of ferric Aplysia limacina myoglobin in different liganded states.
J.Mol.Biol., 233, 1993
|
|
