3OC3
 
 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
2A3Y
 
 | | Pentameric crystal structure of human serum amyloid P-component bound to Bis-1,2-{[(Z)-2carboxy-2-methyl-1,3-dioxane]-5-yloxycarbamoyl}-ethane. | | Descriptor: | BIS-1,2-{[(Z)-2-CARBOXY-2-METHYL-1,3-DIOXANE]-5-YLOXYCARBAMOYL}-ETHANE, CALCIUM ION, Serum amyloid P-component | | Authors: | Ho, J.G, Kitov, P.I, Paszkiewicz, E, Sadowska, J, Bundle, D.R, Ng, K.K. | | Deposit date: | 2005-06-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-assisted Aggregation of Proteins: DIMERIZATION OF SERUM AMYLOID P COMPONENT BY BIVALENT LIGANDS.
J.Biol.Chem., 280, 2005
|
|
1E3X
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.92A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-26 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
5VV4
 
 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 4-(2-(((2-Aminoquinolin-7-yl)methyl)amino)ethyl)-2-methylbenzonitrile | | Descriptor: | 4-(2-{[(2-aminoquinolin-7-yl)methyl]amino}ethyl)-2-methylbenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
2OM7
 
 | | Structural Basis for Interaction of the Ribosome with the Switch Regions of GTP-bound Elongation Factors | | Descriptor: | 16S ribosomal RNA (H5), 30S ribosomal protein S12, 30S ribosomal protein S2, ... | | Authors: | Connell, S.R, Wilson, D.N, Rost, M, Schueler, M, Giesebrecht, J, Dabrowski, M, Mielke, T, Fucini, P, Spahn, C.M.T. | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors.
Mol.Cell, 25, 2007
|
|
3LL8
 
 | |
1M1K
 
 | | Co-crystal structure of azithromycin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, AZITHROMYCIN, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-06-19 | | Release date: | 2002-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
4OSO
 
 | | The crystal structure of landomycin C-6 ketoreductase LanV with bound NADP and rabelomycin | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reductase homolog, ... | | Authors: | Paananen, P, Niiranen, L, Patrikainen, P, Metsa-Ketela, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based engineering of angucyclinone 6-ketoreductases.
Chem.Biol., 21, 2014
|
|
4OTD
 
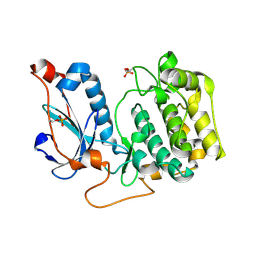 | | Crystal Structure of PRK1 Catalytic Domain | | Descriptor: | Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
4AWI
 
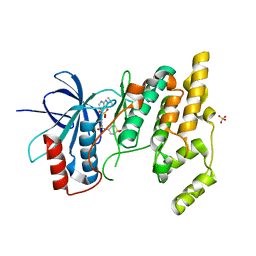 | | Human Jnk1alpha kinase with 4-phenyl-7-azaindole IKK2 inhibitor. | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 8, N-(1,1-dioxidotetrahydro-2H-thiopyran-4-yl)-4-[2-(1-methylethyl)-1H-pyrrolo[2,3-b]pyridin-4-yl]benzenesulfonamide, SULFATE ION | | Authors: | Chung, C, Vicentini, G, Liddle, J, Bamborough, P. | | Deposit date: | 2012-06-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | 4-Phenyl-7-Azaindoles as Potent, Selective and Bioavailable Ikk2 Inhibitors Demonstrating Good in Vivo Efficacy.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1M8W
 
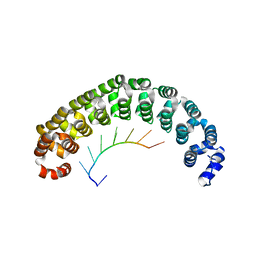 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE1-19 RNA | | Descriptor: | 5'-R(P*UP*GP*UP*AP*UP*AP*U)-3', 5'-R(P*UP*GP*UP*CP*CP*AP*G)-3', 5'-R(P*UP*UP*GP*UP*AP*UP*AP*U)-3', ... | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
3LU7
 
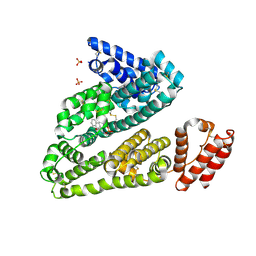 | | Human serum albumin in complex with compound 2 | | Descriptor: | 4-[(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]butanoic acid, PHOSPHATE ION, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
1SL3
 
 | | crystal structue of Thrombin in complex with a potent P1 heterocycle-Aryl based inhibitor | | Descriptor: | (2-[6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDOPYRIDIN-2-YL)ETHYL]AMINO}-2-OXOPYRAZIN-1(2H)-YL]-N-[5-CHLORO-2-(1H-TETRAZOL-1-YL)BENZYL]ACETAMIDE, Hirudin, thrombin | | Authors: | Young, M.B, Barrow, J.C, Glass, K.L, Lundell, G.F, Newton, C.L, Pellicore, J.M, Rittle, K.E, Selnick, H.G, Stauffer, K.J, Vacca, J.P, Williams, P.D, Bohn, D, Clayton, F.C, Cook, J.J, Krueger, J.A, Kuo, L.C, Lewis, S.D, Lucas, B.J, McMasters, D.R, Miller-Stein, C, Pietrak, B.L. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and evaluation of potent P1 aryl heterocycle-based thrombin inhibitors
J.Med.Chem., 47, 2004
|
|
1PP8
 
 | | crystal structure of the T. vaginalis IBP39 Initiator binding domain (IBD) bound to the alpha-SCS Inr element | | Descriptor: | 39 kDa initiator binding protein, ALPHA-SCS INR, SULFATE ION | | Authors: | Schumacher, M.A, Lau, A.O.T, Johnson, P.J. | | Deposit date: | 2003-06-16 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Basis of Core Promoter Recognition in a Primitive Eukaryote
Cell(Cambridge,Mass.), 115, 2003
|
|
2A3X
 
 | | Decameric crystal structure of human serum amyloid P-component bound to Bis-1,2-{[(Z)-2carboxy- 2-methyl-1,3-dioxane]- 5-yloxycarbonyl}-piperazine | | Descriptor: | BIS-1,2-{[(Z)-2CARBOXY-2-METHYL-1,3-DIOXANE]-5-YLOXYCARBONYL}-PIPERAZINE, CALCIUM ION, Serum amyloid P-component | | Authors: | Ho, J.G, Kitov, P.I, Paszkiewicz, E, Sadowska, J, Bundle, D.R, Ng, K.K. | | Deposit date: | 2005-06-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ligand-assisted Aggregation of Proteins: DIMERIZATION OF SERUM AMYLOID P COMPONENT BY BIVALENT LIGANDS.
J.Biol.Chem., 280, 2005
|
|
4C35
 
 | | PKA-S6K1 Chimera with compound 1 (NU1085) bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-hydroxyphenyl)-1H-benzimidazole-4-carboxamide, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
5REB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434899 | | Descriptor: | 1-[(thiophen-3-yl)methyl]piperidin-4-ol, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5W62
 
 | | Crystal structure of mouse BAX monomer | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
5RGO
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102248 (Mpro-x0736) | | Descriptor: | 1-[4-(furan-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2YKA
 
 | | RRM domain of mRNA export adaptor REF2-I bound to HVS ORF57 peptide | | Descriptor: | 52 KDA IMMEDIATE-EARLY PHOSPHOPROTEIN, RNA AND EXPORT FACTOR-BINDING PROTEIN 2 | | Authors: | Tunnicliffe, R.B, Hautbergue, G.M, Kalra, P, Wilson, S.A, Golovanov, A.P. | | Deposit date: | 2011-05-26 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri Orf57 to the Mammalian Export Adaptor Alyref.
Plos Pathog., 10, 2014
|
|
5RF2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1741969146 | | Descriptor: | 1-azanylpropylideneazanium, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102372 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(3S)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-phenylacetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102121 | | Descriptor: | 1-{4-[(5-chlorothiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4BWB
 
 | | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN, NEUROTENSIN RECEPTOR TYPE 1 | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-07-01 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
