7NNY
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with naphthalen-1-ol. | | Descriptor: | 1-NAPHTHOL, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
5IGM
 
 | | Crystal structure of the bromodomain of human BRD9 in complex with bromosporine (BSP) | | Descriptor: | Bromodomain-containing protein 9, Bromosporine | | Authors: | Tallant, C, Filippakopoulos, P, Picaud, S, Nunez-Alonso, G, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
7NPH
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with 5-methoxy-1,3-benzoxazole-2-carboxylic acid | | Descriptor: | 5-methoxy-1,3-benzoxazole-2-carboxylic acid, N-acetyl-gamma-glutamyl-phosphate reductase, PHOSPHATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
5LRN
 
 | | Structure of mono-zinc MCR-1 in P21 space group | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hinchliffe, P, Paterson, N.G, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
7NNZ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 5-methyl-4-phenylthiazol-2-amine. | | Descriptor: | 5-methyl-4-phenyl-1,3-thiazol-2-amine, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4X8P
 
 | | Crystal structure of Ash2L SPRY domain in complex with RbBP5 | | Descriptor: | GLYCEROL, Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2,Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
7POT
 
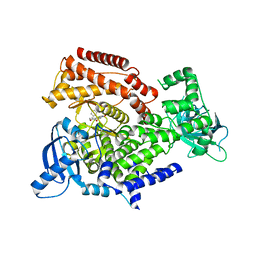 | | PI3 kinase delta in complex with N-[5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxypyridin-3-yl]benzenesulfonamide | | Descriptor: | N-[5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxy-pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
5II2
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CITRIC ACID, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
7POR
 
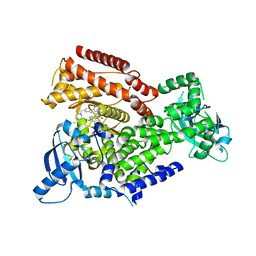 | |
6S3A
 
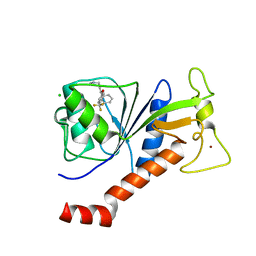 | | Coxsackie B3 2C protein in complex with S-Fluoxetine | | Descriptor: | (3S)-N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 2C protein, CHLORIDE ION, ... | | Authors: | El Kazzi, P, Papageorgiou, N, Ferron, F.P, Bauer, L, van Kuppeveld, F, Coutard, B. | | Deposit date: | 2019-06-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes a ring-shaped hexameric complex.
Sci Adv, 8, 2022
|
|
7POS
 
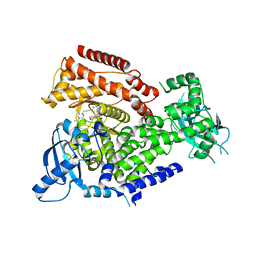 | | PI3 kinase delta in complex with 5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxy-N-(5-{3-[4-(propan-2-yl)piperazin-1-yl]prop-1-yn-1-yl}pyridin-3-yl)pyridine-3-sulfonamide | | Descriptor: | 5-(3,6-dihydro-2~{H}-pyran-4-yl)-2-methoxy-~{N}-[5-[3-(4-propan-2-ylpiperazin-1-yl)prop-1-ynyl]pyridin-3-yl]pyridine-3-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
7POP
 
 | | PI3 kinase delta in complex with 5-[3,6-dihydro-2H-pyran-4-yl]-2-methoxy-N-[2-methylpyridin-4-yl]pyridine-3-sulfonamide | | Descriptor: | 5-[3,6-dihydro-2H-pyran-4-yl]-2-methoxy-N-[2-methylpyridin-4-yl]pyridine-3-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
7NOR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 2-fluoro-4-hydroxybenzonitrile. | | Descriptor: | 2-fluoro-4-hydroxybenzonitrile, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NOV
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (4-methyl-3-nitrophenyl)boronic acid. | | Descriptor: | (4-methyl-3-nitro-phenyl)-oxidanyl-oxidanylidene-boron, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with methyl 4-hydroxy-3-iodobenzoate. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION, methyl 3-iodanyl-4-oxidanyl-benzoate | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8TXH
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 14 | | Descriptor: | (4P)-2-amino-4-{4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}-6-(trifluoromethyl)quinazolin-7-yl}-7-fluoro-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
7N3D
 
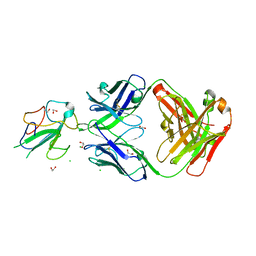 | | Crystal Structure of Human Fab S24-1564 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nucleoprotein, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
8TXE
 
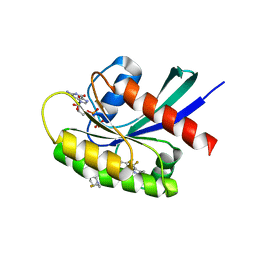 | | Crystal structure of KRAS G12D in complex with GDP and compound 5 | | Descriptor: | (6M)-6-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-4-methyl-5-(trifluoromethyl)pyridin-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
6S6C
 
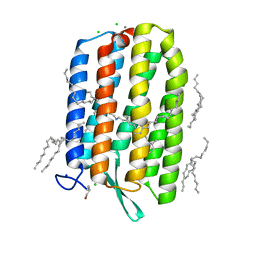 | | Ground state structure of Archaerhodopsin-3 at 100K | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Axford, D, Kwan, T.O.C, Bada Juarez, J.F, Vinals, J, Watts, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structures of the archaerhodopsin-3 transporter reveal that disordering of internal water networks underpins receptor sensitization.
Nat Commun, 12, 2021
|
|
8FWI
 
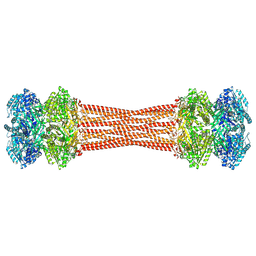 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
7AQB
 
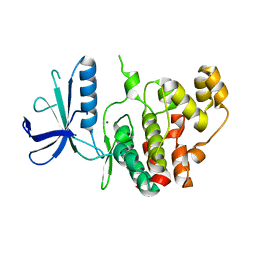 | | Crystal structure of human mitogen activated protein kinase 6 (MAPK6) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mitogen-activated protein kinase 6 | | Authors: | Filippakopoulos, P, Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure and Inhibitor Identifications Reveal Targeting Opportunity for the Atypical MAPK Kinase ERK3.
Int J Mol Sci, 21, 2020
|
|
7NJ1
 
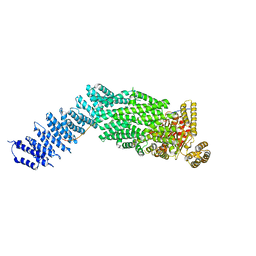 | | CryoEM structure of the human Separase-Securin complex | | Descriptor: | Securin, Separin | | Authors: | Yu, J, Raia, P, Ghent, C.M, Raisch, T, Sadian, Y, Barford, D, Raunser, S, Morgan, D.O, Boland, A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human separase regulation by securin and CDK1-cyclin B1.
Nature, 596, 2021
|
|
4X18
 
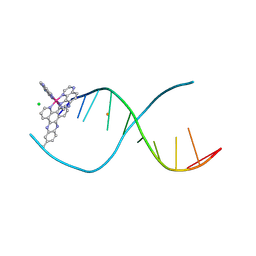 | | [Ru(TAP)2(dppz-11-Me)]2+ bound to d(TCGGCGCCGA) | | Descriptor: | (11-methyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium, BARIUM ION, CHLORIDE ION, ... | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-11-24 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The Structural Effect of Methyl Substitution on the Binding of Polypyridyl Ru-dppz Complexes to DNA
Organometallics, 2015
|
|
4X1A
 
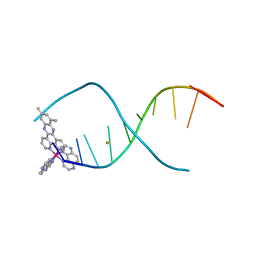 | | Lambda-[Ru(TAP)2(dppz-10,12-Me)]2+ bound to d(TCGGCGCCGA) | | Descriptor: | (10,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium, BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3') | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-11-24 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The Structural Effect of Methyl Substitution on the Binding of Polypyridyl Ru-dppz Complexes to DNA
Organometallics, 2015
|
|
