2G0V
 
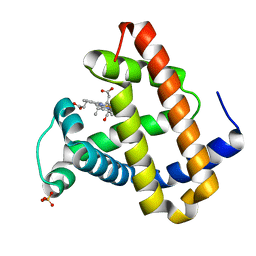 | | Photolyzed CO L29F Myoglobin: 100ps | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Aranda, R, Levin, E.J, Schotte, F, Anfinrud, P.A, Phillips Jr, G.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7YFJ
 
 | | Crystal structure of human WTAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pre-mRNA-splicing regulator WTAP | | Authors: | Yan, X.H, Guan, Z.Y, Tang, C, Yin, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AI-empowered integrative structural characterization of m 6 A methyltransferase complex.
Cell Res., 32, 2022
|
|
3HF4
 
 | |
7YI8
 
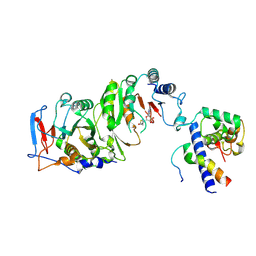 | | Cryo-EM structure of SAH-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
7YI9
 
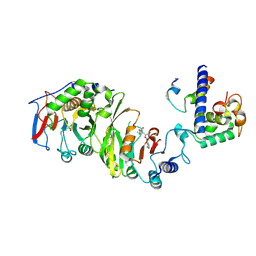 | | Cryo-EM structure of SAM-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
8DNN
 
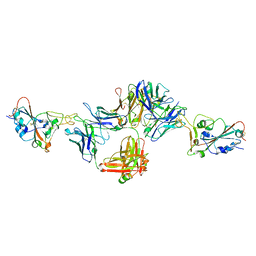 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
2G66
 
 | |
2G6Q
 
 | |
1N1L
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND INHIBITOR (GW472467X) | | Descriptor: | HCV NS3 SERINE PROTEASE, NS4A COFACTOR, ZINC ION, ... | | Authors: | Andrews, D.M, Chaignot, H, Coomber, B.A, Good, A.C, Hind, S.L, Jones, P.S, Mill, G, Robinson, J.E, Skarzynski, T, Slater, M.J, Somers, D.O.N. | | Deposit date: | 2002-10-18 | | Release date: | 2003-10-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 2. The use of X-ray Crystal Structure Data in the Optimisation of P3 and P4 Substituents
Org.Lett., 4, 2002
|
|
2G7N
 
 | | Structure of the Light Chain of Botulinum Neurotoxin Serotype A Bound to small Molecule Inhibitors | | Descriptor: | Botulinum neurotoxin type A, SILVER ION, ZINC ION | | Authors: | Fu, Z, Baldwin, M.R, Boldt, G.E, Janda, K.D, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Light Chain of Botulinum Neurotoxin Serotype A bound to Small Molecule Inhibitors
To be Published
|
|
2GGN
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
2FZC
 
 | | The Structure of Wild-Type E. Coli Aspartate Transcarbamoylase in Complex with Novel T State Inhibitors at 2.10 Resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heng, S, Stieglitz, K.A, Eldo, J, Xia, J, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | T-state Inhibitors of E. coli Aspartate Transcarbamoylase that Prevent the Allosteric Transition.
Biochemistry, 45, 2006
|
|
3UVM
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL4 | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
1N28
 
 | | Crystal structure of the H48Q mutant of human group IIA phospholipase A2 | | Descriptor: | CALCIUM ION, Phospholipase A2, membrane associated | | Authors: | Edwards, S.H, Thompson, D, Baker, S.F, Wood, S.P, Wilton, D.C. | | Deposit date: | 2002-10-22 | | Release date: | 2003-10-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism
Biochemistry, 41, 2002
|
|
2G0X
 
 | | Photolyzed CO L29F Myoglobin: 316ps | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Aranda, R, Levin, E.J, Schotte, F, Anfinrud, P.A, Phillips Jr, G.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1N19
 
 | | Structure of the HSOD A4V mutant | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide Dismutase [Cu-Zn], ... | | Authors: | Cardoso, R.M.F, Thayer, M.M, DiDonato, M, Lo, T.P, Bruns, C.K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2002-10-16 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase.
J.Mol.Biol., 324, 2002
|
|
8DP1
 
 | | Cryo-EM structure of HIV-1 Env(BG505.T332N SOSIP) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
2G14
 
 | | Photolyzed CO L29F Myoglobin: 3.16us | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Aranda, R, Levin, E.J, Schotte, F, Anfinrud, P.A, Phillips Jr, G.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
8DNQ
 
 | | BRD2-BD1 in complex with cyclic peptide 2.2B | | Descriptor: | Bromodomain-containing protein 2, Cyclic peptide 2.2B, GLYCEROL | | Authors: | Patel, K, Franck, C, Mackay, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
2G60
 
 | | Structure of anti-FLAG M2 Fab domain | | Descriptor: | anti-FLAG M2 Fab heavy chain, anti-FLAG M2 Fab light chain | | Authors: | Roosild, T.P. | | Deposit date: | 2006-02-23 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of anti-FLAG M2 Fab domain and its use in the stabilization of engineered membrane proteins.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GHK
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
8DOW
 
 | | Cryo-EM structure of HIV-1 Env(CH848 10.17 DS.SOSIP_DT) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
8DWO
 
 | | Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab | | Descriptor: | Envelope glycoprotein E1, Envelope glycoprotein E2, SKE26 Fab Heavy Chain, ... | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
2G9W
 
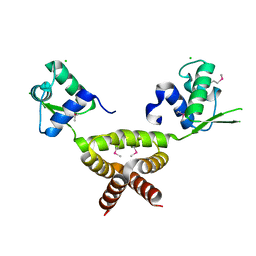 | | Crystal Structure of Rv1846c, a Putative Transcriptional Regulatory Protein of Mycobacterium Tuberculosis | | Descriptor: | CHLORIDE ION, conserved hypothetical protein | | Authors: | Saul, F.A, Haouz, A, Fiez-Vandal, C, Shepard, W, Alzari, P.M. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis
Mol.Microbiol., 71, 2009
|
|
2G93
 
 | |
