5EGS
 
 | | Human PRMT6 with bound fragment-type inhibitor | | Descriptor: | 2-[4-(phenylmethyl)piperidin-1-yl]ethanamine, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Steuber, H, Egner, U, Kania, J, Wu, H, Brown, P.J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Potent Class I Protein Arginine Methyltransferase Fragment Inhibitor.
J.Med.Chem., 59, 2016
|
|
2WW5
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | Deposit date: | 2009-10-21 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I5K
 
 | | Crystal structure of the NS5B polymerase from Hepatitis C Virus (HCV) strain JFH1 | | Descriptor: | PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Simister, P.C, Schmitt, M, Lohmann, V, Bressanelli, S. | | Deposit date: | 2009-07-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of hepatitis C virus strain JFH1 polymerase
J.Virol., 83, 2009
|
|
5EHC
 
 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-chlorophenyl)methyl]-6-oxidanylidene-1~{H}-purin-7-ium-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-4-oxidanyl-cyclobut-3-ene-1,2-dione, Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
5NUN
 
 | | Engineered beta-lactoglobulin: variant F105L-L39A-M107F in complex with chlorpromazine (LG-LAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Towrzydlo, M, Lazinskia, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
4X3F
 
 | |
5NZS
 
 | | The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor GTPase-activating protein 2, Coatomer subunit alpha, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5ULG
 
 | |
1LGV
 
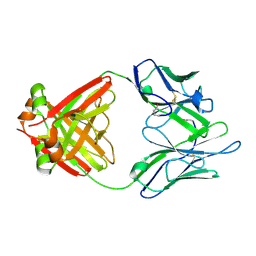 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 100K | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | Deposit date: | 2002-04-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
8VT3
 
 | | cryo-EM structure of HMPV (MPV-2cREKR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prefusion of HMPV (MPV-2cREKR), ... | | Authors: | Yu, X, Langedijk, J.P.M. | | Deposit date: | 2024-01-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer.
Nat Commun, 15, 2024
|
|
1OXL
 
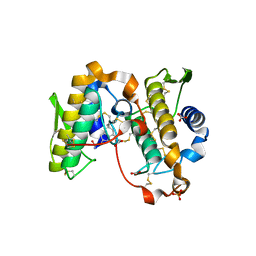 | | INHIBITION OF PHOSPHOLIPASE A2 (PLA2) BY (2-CARBAMOYLMETHYL-5-PROPYL-OCTAHYDRO-INDOL-7-YL)-ACETIC ACID (INDOLE): CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PLA2 FROM RUSSELL'S VIPER AND INDOLE AT 1.8 RESOLUTION | | Descriptor: | (2-CARBAMOYLMETHYL-5-PROPYL-OCTAHYDRO-INDOL-7-YL)ACETIC ACID, CARBONATE ION, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Chandra, V, Balasubramanya, R, Kaur, P, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of the secretory phospholipase A2 from Daboia russelli pulchella with an endogenic indole derivative, 2-carbamoylmethyl-5-propyl-octahydro-indol-7-yl-acetic acid at 1.8 A resolution.
Biochim.Biophys.Acta, 1752, 2005
|
|
5UNQ
 
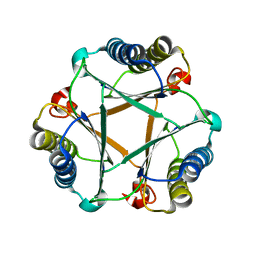 | |
1P4E
 
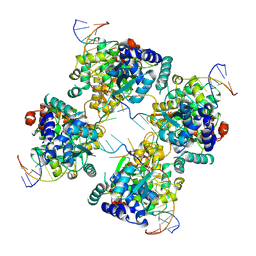 | | Flpe W330F mutant-DNA Holliday Junction Complex | | Descriptor: | 33-MER, 5'-D(*TP*AP*AP*GP*TP*TP*CP*CP*TP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*TP*AP*GP*GP*AP*AP*CP*TP*TP*C)-3', ... | | Authors: | Rice, P.A, Chen, Y. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis
J.Biol.Chem., 278, 2003
|
|
4WST
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
6NTI
 
 | | Neutron/X-ray crystal structure of AAC-VIa bound to kanamycin b | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5LUR
 
 | | An avidin mutant | | Descriptor: | Avidin, CHLORIDE ION, PROGESTERONE | | Authors: | Agrawal, N, Lehtonen, S.I, Riihimaki, T.A, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An avidin mutant
TO BE PUBLISHED
|
|
8WCH
 
 | |
4YXM
 
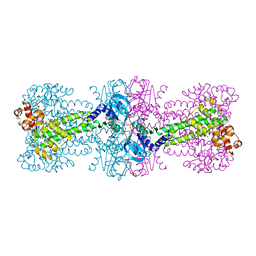 | | Structure of Thermotoga maritima DisA D75N mutant with reaction product c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA integrity scanning protein DisA | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
7LNW
 
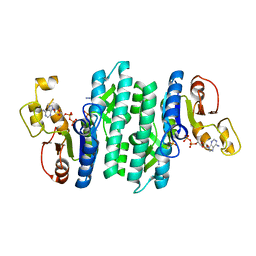 | |
7LNX
 
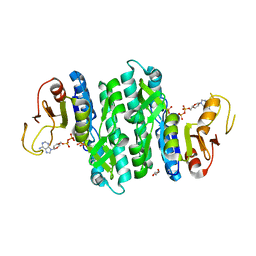 | | I146A mutant of the isopentenyl phosphate kinase from Candidatus methanomethylophilus alvus | | Descriptor: | (2E)-3-methylhept-2-en-1-yl dihydrogen phosphate, (2Z)-3-methylhept-2-en-1-yl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Johnson, B.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
1P5G
 
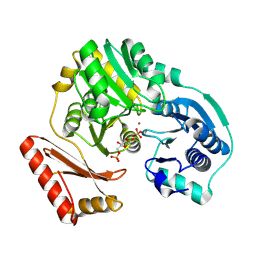 | |
2GL1
 
 | | NMR solution structure of Vigna radiata Defensin 2 (VrD2) | | Descriptor: | PDF1 | | Authors: | Lin, K.F, Lee, T.R, Tsai, P.H, Hsu, M.P, Chen, C.S, Lyu, P.C. | | Deposit date: | 2006-04-04 | | Release date: | 2007-04-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-based protein engineering for alpha-amylase inhibitory activity of plant defensin.
Proteins, 68, 2007
|
|
5LIW
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK319 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5G3L
 
 | | ESCHERICHIA COLI HEAT LABILE ENTEROTOXIN TYPE IIB B-PENTAMER COMPLEXED WITH SIALYLATED SUGAR | | Descriptor: | HEAT-LABILE ENTEROTOXIN IIB, B CHAIN, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Zalem, D, Benktander, J, Ribeiro, J.P, Varrot, A, Lebens, M, Imberty, A, Teneberg, S. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Biochemical and Structural Characterization of the Novel Sialic Acid-Binding Site of Escherichia Coli Heat-Labile Enterotoxin Lt-Iib.
Biochem.J., 473, 2016
|
|
1H79
 
 | | STRUCTURAL BASIS FOR ALLOSTERIC SUBSTRATE SPECIFICITY REGULATION IN CLASS III RIBONUCLEOTIDE REDUCTASES: NRDD IN COMPLEX WITH DTTP | | Descriptor: | ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2001-07-04 | | Release date: | 2002-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
