7R4N
 
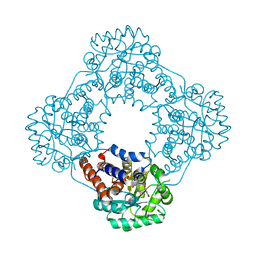 | | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-methyl-1H-indazole-3-carboxamide, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
6NWD
 
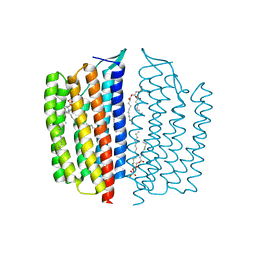 | | X-ray Crystallographic structure of Gloeobacter rhodopsin | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DECANE, DODECANE, ... | | Authors: | Ernst, O.P, Morizumi, T, Ou, W.L. | | Deposit date: | 2019-02-06 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Crystallographic Structure and Oligomerization of Gloeobacter Rhodopsin.
Sci Rep, 9, 2019
|
|
7RHQ
 
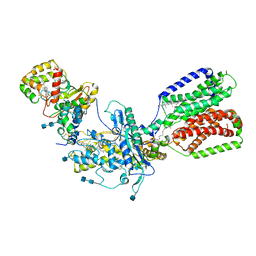 | | Cryo-EM structure of Xenopus Patched-1 in complex with GAS1 and Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, P, Lian, T, Wierbowski, B, Garcia-Linares, S, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
7R70
 
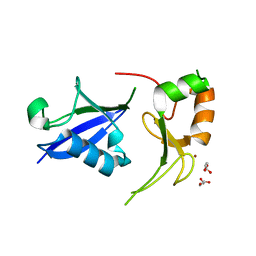 | | Crystal Structure of the UbArk2C fusion protein | | Descriptor: | GLYCEROL, Ubiquitin,E3 ubiquitin-protein ligase RNF165, ZINC ION | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
6NWT
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
7R71
 
 | | Crystal Structure of the UbArk2C-UbcH5b~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin,E3 ubiquitin-protein ligase RNF165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
8PM6
 
 | | Human bile salt export pump (BSEP) in complex with inhibitor GBM in nanodiscs | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Bile salt export pump, CHOLESTEROL | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
6NXW
 
 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218S mutant | | Descriptor: | Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2017-12-08 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
8QKK
 
 | | Cryo-EM structure of MmpL3 from Mycobacterium smegmatis reconstituted into peptidiscs | | Descriptor: | Trehalose monomycolate exporter MmpL3 | | Authors: | Couston, J, Guo, Z, Wang, K, Gourdon, P.E, Blaise, M. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of the trehalose monomycolate transporter, MmpL3, reconstituted into peptidiscs.
Curr Res Struct Biol, 6, 2023
|
|
5ZNC
 
 | |
7B5P
 
 | | AcrB in cycloalkane amphipol | | Descriptor: | Efflux pump membrane transporter | | Authors: | Higgins, A.J, Flynn, A.J, Muench, S.P. | | Deposit date: | 2020-12-05 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cycloalkane-modified amphiphilic polymers provide direct extraction of membrane proteins for CryoEM analysis.
Commun Biol, 4, 2021
|
|
3K22
 
 | | Glucocorticoid Receptor with Bound alaninamide 10 with TIF2 peptide | | Descriptor: | Glucocorticoid receptor, N-[(1R)-2-amino-1-methyl-2-oxoethyl]-3-(6-methyl-4-{[3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propyl]amino}-1H-indazol-1-yl)benzamide, Transcriptional Intermediary Factor 2, ... | | Authors: | Biggadike, K.B, McLay, I.M, Madauss, K.P, Williams, S.P, Bledsoe, R.K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-08-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6OJW
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 holo-LsdA | | Descriptor: | FE (III) ION, GLYCEROL, Lignostilbene-alpha,beta-dioxygenase isozyme I, ... | | Authors: | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
1OAO
 
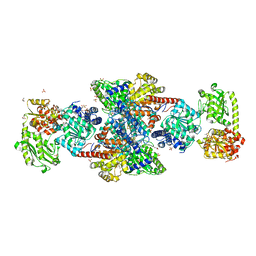 | | NiZn[Fe4S4] and NiNi[Fe4S4] clusters in closed and open alpha subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase | | Descriptor: | ACETATE ION, BICARBONATE ION, CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Darnault, C, Volbeda, A, Kim, E.J, Legrand, P, Vernede, X, Lindahl, P.A, Fontecilla-Camps, J.C. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] Clusters in Closed and Open Alpha Subunits of Acetyl-Coa Synthase/Carbon Monoxide Dehydrogenase
Nat.Struct.Biol., 10, 2003
|
|
8PMJ
 
 | | Vanadate-trapped BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, ... | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | Descriptor: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | Authors: | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
7LHS
 
 | |
6O5D
 
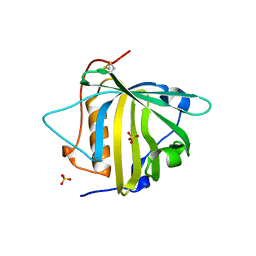 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
3N3M
 
 | |
7MC1
 
 | | Solution structure of Miz-1 Zinc finger 10 | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
4NBT
 
 | | Crystal structure of FabG from Acholeplasma laidlawii | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
7MC2
 
 | | Solution structure of Miz-1 Zinc finger 11 H586Y | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7Y39
 
 | | Ubiquitin-like domain of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Draczkowski, P, Hsu, S.T.D. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insight into ZFAND1 and p97 Interaction
To Be Published
|
|
