6RSE
 
 | |
1OCI
 
 | | [3.2.0]bcANA:DNA | | Descriptor: | 5'-D(*CP*TP*GP*A TLBP*AP*TP*GP*CP)-3', 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*GP)-3' | | Authors: | Tommerholt, H.V, Christensen, N.K, Nielsen, P, Wengel, J, Stein, P.C, Jacobsen, J.P, Petersen, M. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a DsDNA Containing a Bicyclic D-Arabino-Configured Nucleotide Fixed in an O4'-Endo Sugar Conformation
Org.Biomol.Chem., 1, 2003
|
|
1AXB
 
 | |
4R7D
 
 | | Fab Hu 15C1 | | Descriptor: | Fab Hu 15C1 Heavy chain, Fab Hu 15C1 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
6CFQ
 
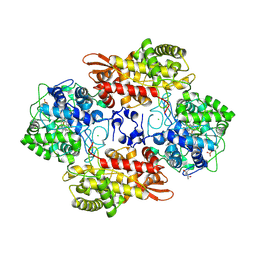 | |
5L4J
 
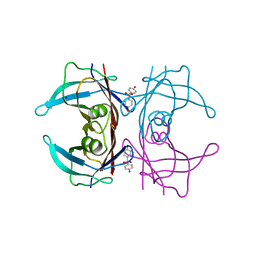 | | Crystal Structure of Human Transthyretin in Complex with 4,4'-Dihydroxydiphenyl sulfone (Bisphenol S, BPS) | | Descriptor: | 4-(4-hydroxyphenyl)sulfonylphenol, SODIUM ION, Transthyretin | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
8AJJ
 
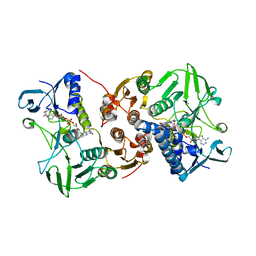 | |
6CD2
 
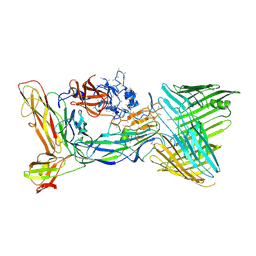 | | Crystal structure of the PapC usher bound to the chaperone-adhesin PapD-PapG | | Descriptor: | Chaperone protein PapD, Outer membrane usher protein PapC, PapGII adhesin protein | | Authors: | Omattage, N.S, Deng, Z, Yuan, P, Hultgren, S.J. | | Deposit date: | 2018-02-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli.
Nat Microbiol, 3, 2018
|
|
2VD6
 
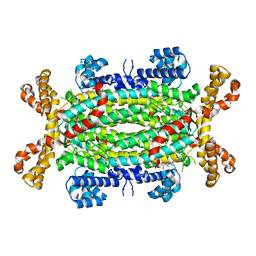 | | Human adenylosuccinate lyase in complex with its substrate N6-(1,2- Dicarboxyethyl)-AMP, and its products AMP and fumarate. | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE LYASE, ... | | Authors: | Stenmark, P, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human Adenylosuccinate Lyase in Complex with its Substrate N6-(1,2-Dicarboxyethyl)-AMP, and its Products AMP and Fumarate.
To be Published
|
|
5UOT
 
 | |
2VJ3
 
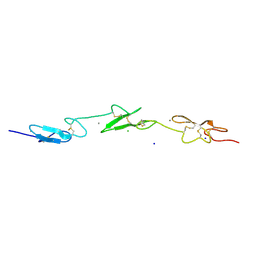 | | Human Notch-1 EGFs 11-13 | | Descriptor: | CALCIUM ION, CHLORIDE ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, ... | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6DVR
 
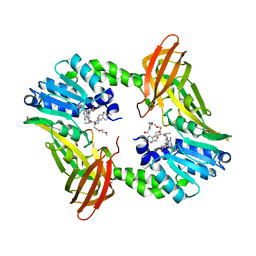 | | Crystal structure of human CARM1 with (R)-SKI-72 | | Descriptor: | (2R,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-methoxyphenyl)ethyl]hexanamide (non-preferred name), 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Zeng, H, Hutchinson, A, Seitova, A, Luo, M, Cai, X.C, Ke, W, Wang, J, Shi, C, Zheng, W, Lee, J.P, Ibanez, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human CARM1 with (R)-SKI-72
to be published
|
|
5IQ7
 
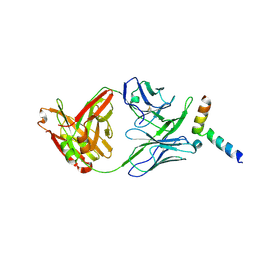 | | Crystal structure of 10E8-S74W Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8-S74W Heavy Chain, 10E8-S74W Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2869 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
5L7Q
 
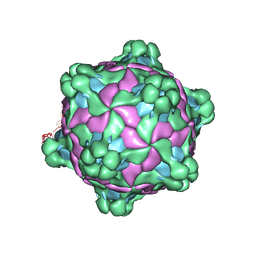 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | VP1, vp2, vp3 | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LB5
 
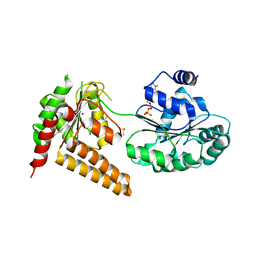 | | Crystal structure of human RECQL5 helicase in complex with ADP/Mg (tricilinc form). | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, DIMETHYL SULFOXIDE, ... | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
6A2N
 
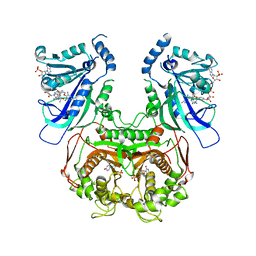 | | Crystal structure of quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum DHFR-TS complexed with BT2, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Tarnchampoo, B, Yuthavong, Y. | | Deposit date: | 2018-06-12 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
8PMR
 
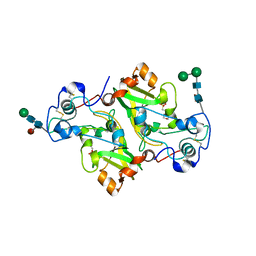 | | NADase from Aspergillus fumigatus with mutated calcium binding motif (D219A/E220A) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
6DVH
 
 | |
6QSW
 
 | | Complement factor B protease domain in complex with the reversible inhibitor N-(2-bromo-4-methylnaphthalen-1-yl)-4,5-dihydro-1H-imidazol-2-amine. | | Descriptor: | Complement factor B, SULFATE ION, ~{N}-(2-bromanyl-4-methyl-naphthalen-1-yl)-4,5-dihydro-1~{H}-imidazol-2-amine | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4R6S
 
 | | Crystal structure of PPARgammma in complex with SR1663 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1R)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
5URF
 
 | | The structure of human bocavirus 1 | | Descriptor: | viral protein 3 | | Authors: | Mietzsch, M, Kailasan, S, Garrison, J, Ilyas, M, Chipman, P, Kandola, K, Jansen, M, Spear, J, Sousa, D, McKenna, R, Soderlund-Venermo, M, Baker, T, Agbandje-McKenna, M. | | Deposit date: | 2017-02-10 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insights into Human Bocaparvoviruses.
J. Virol., 91, 2017
|
|
5DSC
 
 | |
6A8N
 
 | | The crystal structure of muPAin-1-IG-2 in complex with muPA-SPD at pH8.5 | | Descriptor: | CYS-PRO-ALA-TYR-SER-ARG-TYR-ILE-GLY-CYS, Urokinase-type plasminogen activator B | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
5JEO
 
 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
