3ZUK
 
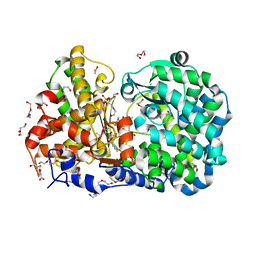 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE ZMP1 IN COMPLEX WITH INHIBITOR | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Ferraris, D.M, Sbardella, D, Petrera, A, Marini, S, Amstutz, B, Coletta, M, Sander, P, Rizzi, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Zinc-Dependent Metalloprotease-1 (Zmp1), a Metalloprotease Involved in Pathogenicity.
J.Biol.Chem., 286, 2011
|
|
4R2U
 
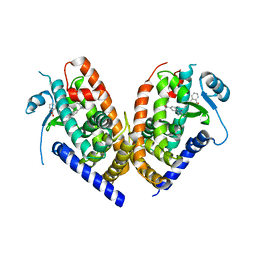 | | Crystal Structure of PPARgamma in complex with SR1664 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1S)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Kamenecka, T, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-13 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
2OC3
 
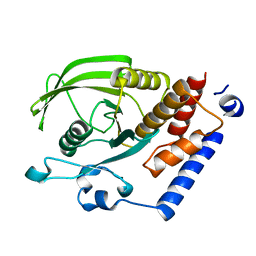 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 18 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Gorrec, F, Umeano, C, Savitsky, P, Sobott, F, Eswaran, J, Papagrigoriou, E, Debreczeni, J.E, Turnbull, A, Bunkoczi, G, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
5A1L
 
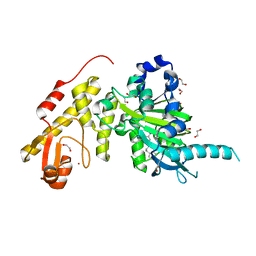 | | Crystal structure of JmjC domain of human histone demethylase UTY with S21056a | | Descriptor: | 1,2-ETHANEDIOL, 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propan-1-ol, FE (II) ION, ... | | Authors: | Srikannathasan, V, Gileadi, C, Johansson, C, Krojer, T, Tumber, A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Oppermann, U. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty with S21056A
To be Published
|
|
2OE2
 
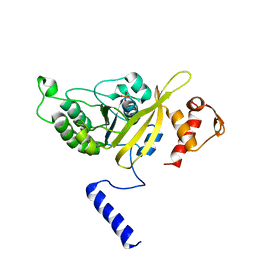 | | MSrecA-native-low humidity 95% | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
8QQ3
 
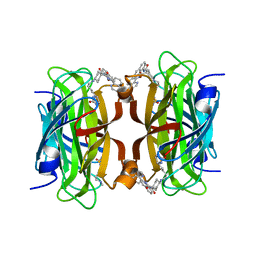 | | Streptavidin with a Ni-cofactor | | Descriptor: | 4-[4-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butylamino]-~{N}1,~{N}1'-di(quinolin-8-yl)cyclohexane-1,1-dicarboxamide, NICKEL (II) ION, Streptavidin | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial nickel chlorinase based on the biotin-streptavidin technology.
Chem.Commun.(Camb.), 60, 2024
|
|
2WE3
 
 | | EBV dUTPase inactive mutant deleted of motif V | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
5UC5
 
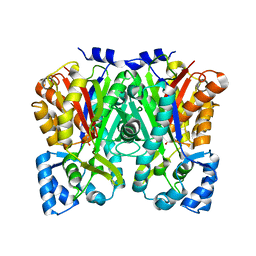 | | Chalcone synthase from Malus domestica | | Descriptor: | CHS2 chalcone synthase | | Authors: | Stewart Jr, C.E, Noel, J.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular architectures of benzoic acid-specific type III polyketide synthases.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NUE
 
 | | Cytosolic Malate Dehydrogenase 1 (peroxide-treated) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ETHOXYETHANOL, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-02-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.35000277 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
1FEQ
 
 | | NMR SOLUTION STRUCTURE OF THE ANTICODON OF TRNA(LYS3) WITH T6A MODIFICATION AT POSITION 37 | | Descriptor: | 5'-R(*GP*CP*AP*GP*AP*CP*UP*UP*UP*UP*(T6A)P*AP*UP*CP*UP*GP*C)-3' | | Authors: | Stuart, J.W, Gdaniec, Z, Guenther, R.H, Marszalek, M, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2000-07-21 | | Release date: | 2000-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A.
Biochemistry, 39, 2000
|
|
4HZE
 
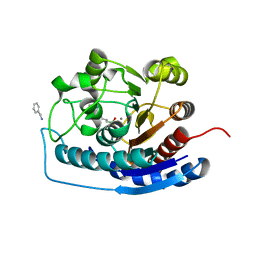 | | Crystal structure of human Arginase-2 complexed with inhibitor 9 | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-15 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
2OGT
 
 | | Crystal Structure of the Geobacillus Stearothermophilus Carboxylesterase EST55 at pH 6.8 | | Descriptor: | GLYCEROL, IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
1O27
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and BrdUMP at 2.3 A resolution | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
8RE3
 
 | | Crystal Structure determination of Dye-decolorizing Peroxidase (DyP) mutant M190G from Deinoccoccus radiodurans | | Descriptor: | CALCIUM ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Salgueiro, B.A, Frade, K, Frazao, C, Matias, P, Moe, E. | | Deposit date: | 2023-12-10 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical, Biophysical, and Structural Analysis of an Unusual DyP from the Extremophile Deinococcus radiodurans.
Molecules, 29, 2024
|
|
5UDR
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with nicotinamide mononucleotid NMN | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MANGANESE (II) ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDV
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with iron | | Descriptor: | FE (III) ION, Lactate racemization operon protein LarE, PHOSPHATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6T9I
 
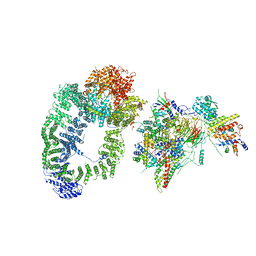 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
3ZSR
 
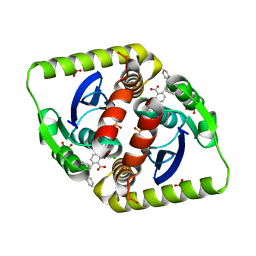 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
5A2F
 
 | | Two membrane distal IgSF domains of CD166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD166 ANTIGEN | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
3ZT4
 
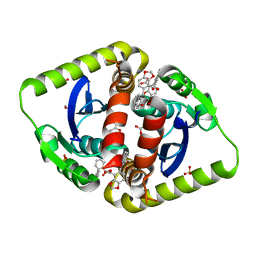 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(2-OXO-2,3-DIHYDRO-1H-INDEN-1-YLIDENE)METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design
Plos One, 7, 2012
|
|
4R20
 
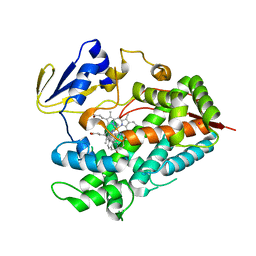 | | Zebra fish cytochrome P450 17A2 with Abiraterone | | Descriptor: | Abiraterone, Cytochrome P450 family 17 polypeptide 2, MERCURY (II) ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural and Kinetic Basis of Steroid 17 alpha, 20-Lyase Activity in Teleost Fish Cytochrome P450 17A1 and Its Absence in Cytochrome P450 17A2.
J.Biol.Chem., 290, 2015
|
|
5NZE
 
 | | Complex of S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Hejdanek, J. | | Deposit date: | 2017-05-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5PG8
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 172) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PGG
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 180) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4I85
 
 | | Crystal structure of transthyretin in complex with CHF5074 at neutral pH | | Descriptor: | 1-(3',4'-dichloro-2-fluorobiphenyl-4-yl)cyclopropanecarboxylic acid, Transthyretin | | Authors: | Zanotti, G, Cendron, L, Folli, C, Florio, P, Imbimbo, B.P, Berni, R. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors.
Febs Lett., 587, 2013
|
|
